Immunity to pathogens is divided into innate and adaptive immune responses. The innate immune response is the 1st line of defense against a variety of pathogens, including bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology, fungi Fungi A kingdom of eukaryotic, heterotrophic organisms that live parasitically as saprobes, including mushrooms; yeasts; smuts, molds, etc. They reproduce either sexually or asexually, and have life cycles that range from simple to complex. Filamentous fungi, commonly known as molds, refer to those that grow as multicellular colonies. Mycology, viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology, and parasites. In essentially the same form, the innate type of immunity is present in all multicellular Multicellular Cell Types: Eukaryotic versus Prokaryotic organisms. The innate immune response is activated within minutes to hours after exposure to an infection, which curtails microbe invasion at the initial stages. The pathogen has specific components recognized by pattern recognition receptors Receptors Receptors are proteins located either on the surface of or within a cell that can bind to signaling molecules known as ligands (e.g., hormones) and cause some type of response within the cell. Receptors (PRRs). After identification Identification Defense Mechanisms of a microbial invasion, noncellular components (including the complement system and cytokines Cytokines Non-antibody proteins secreted by inflammatory leukocytes and some non-leukocytic cells, that act as intercellular mediators. They differ from classical hormones in that they are produced by a number of tissue or cell types rather than by specialized glands. They generally act locally in a paracrine or autocrine rather than endocrine manner. Adaptive Immune Response) act in concert with cellular elements to achieve cell recruitment Recruitment Skeletal Muscle Contraction, direct microbial killing, or phagocytosis Phagocytosis The engulfing and degradation of microorganisms; other cells that are dead, dying, or pathogenic; and foreign particles by phagocytic cells (phagocytes). Innate Immunity: Phagocytes and Antigen Presentation induction. The steps all aim to eliminate the pathogen. Antimicrobial mechanisms in phagocytosis Phagocytosis The engulfing and degradation of microorganisms; other cells that are dead, dying, or pathogenic; and foreign particles by phagocytic cells (phagocytes). Innate Immunity: Phagocytes and Antigen Presentation include acidification and respiratory/oxidative burst. The process terminates with destruction of the threat while maintaining immunologic homeostasis Homeostasis The processes whereby the internal environment of an organism tends to remain balanced and stable. Cell Injury and Death. The defense is also important in activating the adaptive immune system Immune system The body's defense mechanism against foreign organisms or substances and deviant native cells. It includes the humoral immune response and the cell-mediated response and consists of a complex of interrelated cellular, molecular, and genetic components. Primary Lymphatic Organs.
Last updated: Mar 9, 2023
The immune system Immune system The body’s defense mechanism against foreign organisms or substances and deviant native cells. It includes the humoral immune response and the cell-mediated response and consists of a complex of interrelated cellular, molecular, and genetic components. Primary Lymphatic Organs provides defense (immunity) against invading pathogens ranging from viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology to parasites; components are interconnected by blood and lymphatic circulation Circulation The movement of the blood as it is pumped through the cardiovascular system. ABCDE Assessment.
Two lines of overlapping defense:
| Innate immunity Innate immunity The capacity of a normal organism to remain unaffected by microorganisms and their toxins. It results from the presence of naturally occurring anti-infective agents, constitutional factors such as body temperature and immediate acting immune cells such as natural killer cells. Innate Immunity: Phagocytes and Antigen Presentation | Adaptive immunity | |
|---|---|---|
| Genetics Genetics Genetics is the study of genes and their functions and behaviors. Basic Terms of Genetics | Germline encoded | Gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics rearrangements involved in lymphocyte development |
| Immune response | Nonspecific | Highly specific |
| Timing of response | Immediate (minutes to hours) | Develops over a longer period of time |
| Memory Memory Complex mental function having four distinct phases: (1) memorizing or learning, (2) retention, (3) recall, and (4) recognition. Clinically, it is usually subdivided into immediate, recent, and remote memory. Psychiatric Assessment response | None | Responds quickly upon recognition of antigen Antigen Substances that are recognized by the immune system and induce an immune reaction. Vaccination with memory Memory Complex mental function having four distinct phases: (1) memorizing or learning, (2) retention, (3) recall, and (4) recognition. Clinically, it is usually subdivided into immediate, recent, and remote memory. Psychiatric Assessment response |
| Recognition of pathogen | Pattern recognition receptors Receptors Receptors are proteins located either on the surface of or within a cell that can bind to signaling molecules known as ligands (e.g., hormones) and cause some type of response within the cell. Receptors (PRRs) such as toll-like receptors Receptors Receptors are proteins located either on the surface of or within a cell that can bind to signaling molecules known as ligands (e.g., hormones) and cause some type of response within the cell. Receptors (TLRs) recognize pathogen-associated molecular patterns Pathogen-Associated Molecular Patterns Sepsis and Septic Shock ( PAMPs PAMPs Sepsis and Septic Shock) |
|
| Components |
|
|
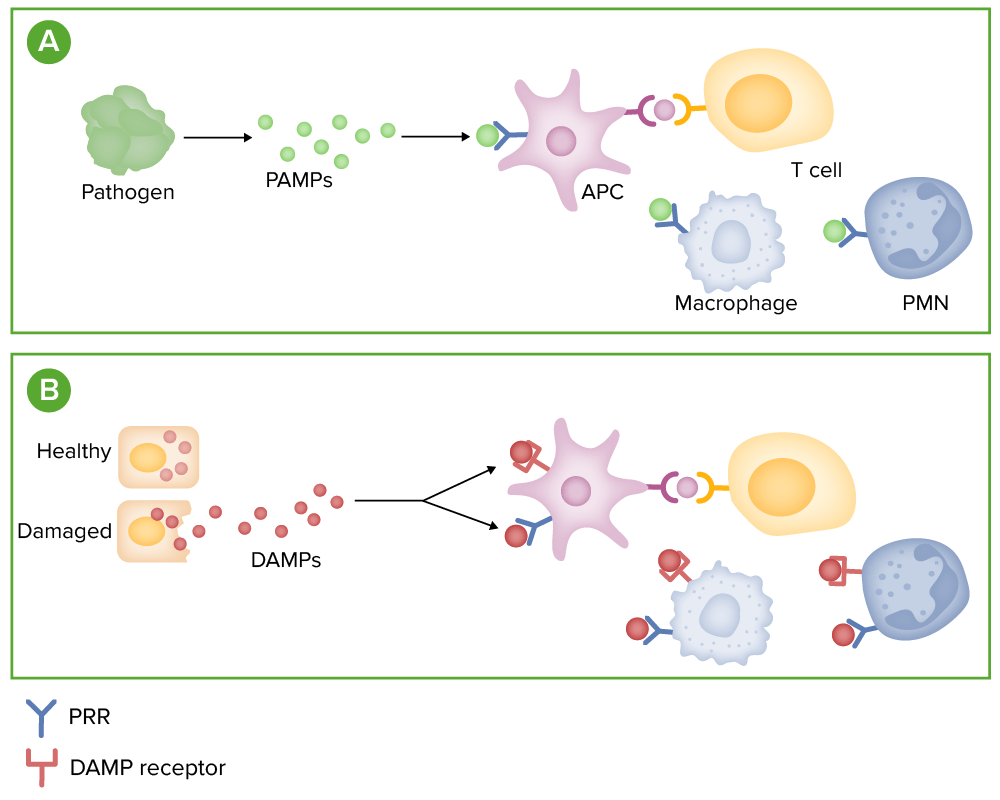
Microbe detection:
(A) Microbes release pathogen-associated molecular patterns (PAMPs), which can bind to pattern recognition receptors (PRRs), such as toll-like receptors, on immune cells.
(B) Stressed or damaged cells release molecules, called damage-associated molecular patterns (DAMPs). These molecules bind to both PRRs and specialied DAMP receptors on immune cells.
Binding of these receptors promotes the release of inflammatory mediators, such as cytokines, chemokines, and complement.
PMN: polymorphonuclear leukocyte
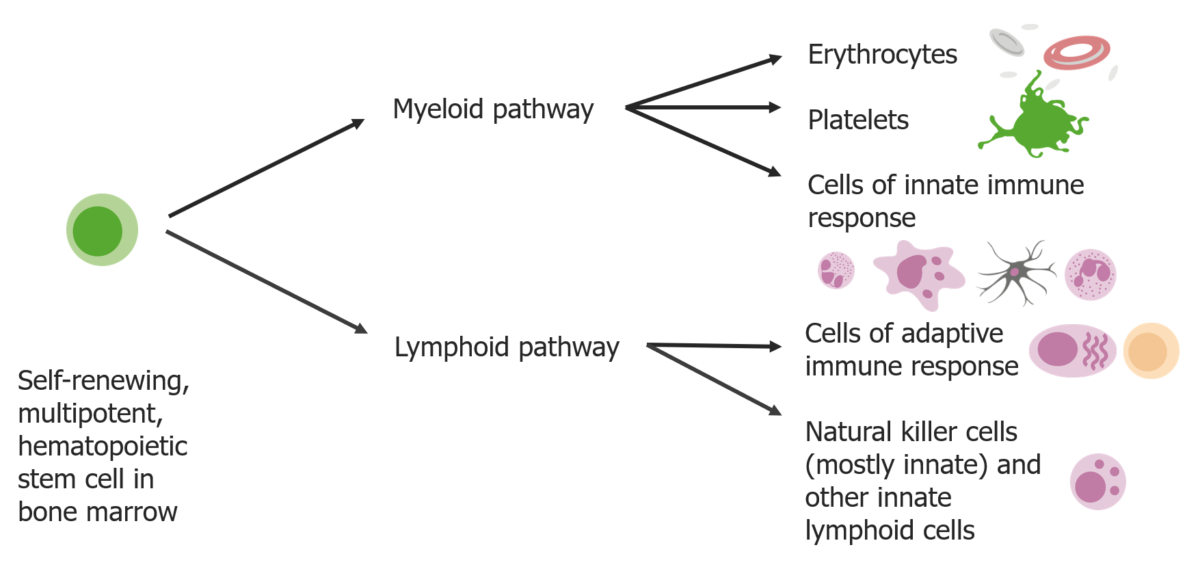
Stem cells differentiate into 2 pathways:
Myeloid pathways produce erythrocytes, platelets, and cells of the innate immune response. Lymphoid pathways produce the cells of adaptive response and natural killer cells.
Epithelial cells line body surfaces and are heavily exposed to antigens.
Includes:
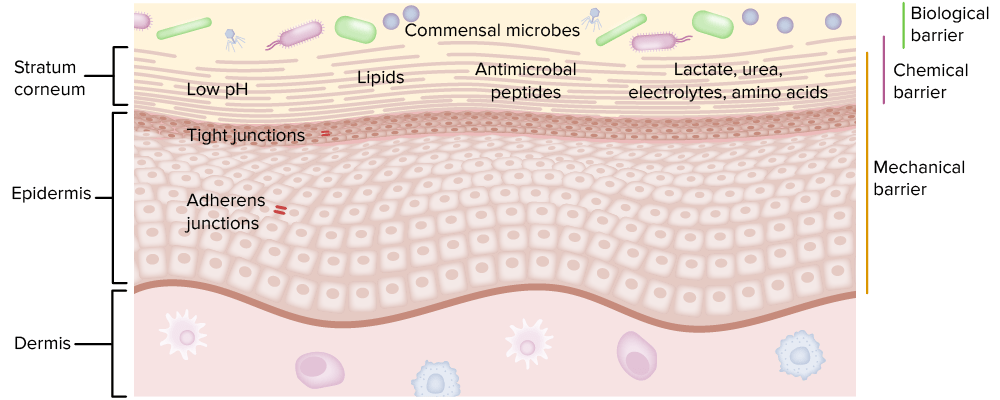
The 3 different types of barriers are an important part of innate immunity.
Image by Lecturio.Cell-associated PRRs are expressed in various immune cells and can be intracellular (endolysosomal/cytoplasmic) or transmembrane.
| Toll-like receptor Receptor Receptors are proteins located either on the surface of or within a cell that can bind to signaling molecules known as ligands (e.g., hormones) and cause some type of response within the cell. Receptors | Localization | Ligand | Origin of the ligand |
|---|---|---|---|
| TLR1 | Plasma membrane Plasma membrane A cell membrane (also known as the plasma membrane or plasmalemma) is a biological membrane that separates the cell contents from the outside environment. A cell membrane is composed of a phospholipid bilayer and proteins that function to protect cellular DNA and mediate the exchange of ions and molecules. The Cell: Cell Membrane | Triacyl lipoprotein | Bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology |
| TLR2 | Lipoprotein | Bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology, viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology, parasites | |
| TLR3 | Endolysosome | dsRNA | Viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology |
| TLR4 | Plasma membrane Plasma membrane A cell membrane (also known as the plasma membrane or plasmalemma) is a biological membrane that separates the cell contents from the outside environment. A cell membrane is composed of a phospholipid bilayer and proteins that function to protect cellular DNA and mediate the exchange of ions and molecules. The Cell: Cell Membrane | LPS | Bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology, viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology |
| TLR5 | Flagellin | Bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology | |
| TLR6 | Diacyl lipoprotein | Bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology, viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology | |
| TLR7, TLR8 | Endolysosome | ssRNA | Viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology, bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology |
| TLR9 | CpG-DNA | Viruses Viruses Minute infectious agents whose genomes are composed of DNA or RNA, but not both. They are characterized by a lack of independent metabolism and the inability to replicate outside living host cells. Virology, bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology, protozoa Protozoa Nitroimidazoles | |
| TLR10 | Unknown | Influenza Influenza Influenza viruses are members of the Orthomyxoviridae family and the causative organisms of influenza, a highly contagious febrile respiratory disease. There are 3 primary influenza viruses (A, B, and C) and various subtypes, which are classified based on their virulent surface antigens, hemagglutinin (HA) and neuraminidase (NA). Influenza typically presents with a fever, myalgia, headache, and symptoms of an upper respiratory infection. Influenza Viruses/Influenza virus Virus Viruses are infectious, obligate intracellular parasites composed of a nucleic acid core surrounded by a protein capsid. Viruses can be either naked (non-enveloped) or enveloped. The classification of viruses is complex and based on many factors, including type and structure of the nucleoid and capsid, the presence of an envelope, the replication cycle, and the host range. Virology, Listeria Listeria Listeria spp. are motile, flagellated, gram-positive, facultative intracellular bacilli. The major pathogenic species is Listeria monocytogenes. Listeria are part of the normal gastrointestinal flora of domestic mammals and poultry and are transmitted to humans through the ingestion of contaminated food, especially unpasteurized dairy products. Listeria Monocytogenes/Listeriosis monocytogenes |
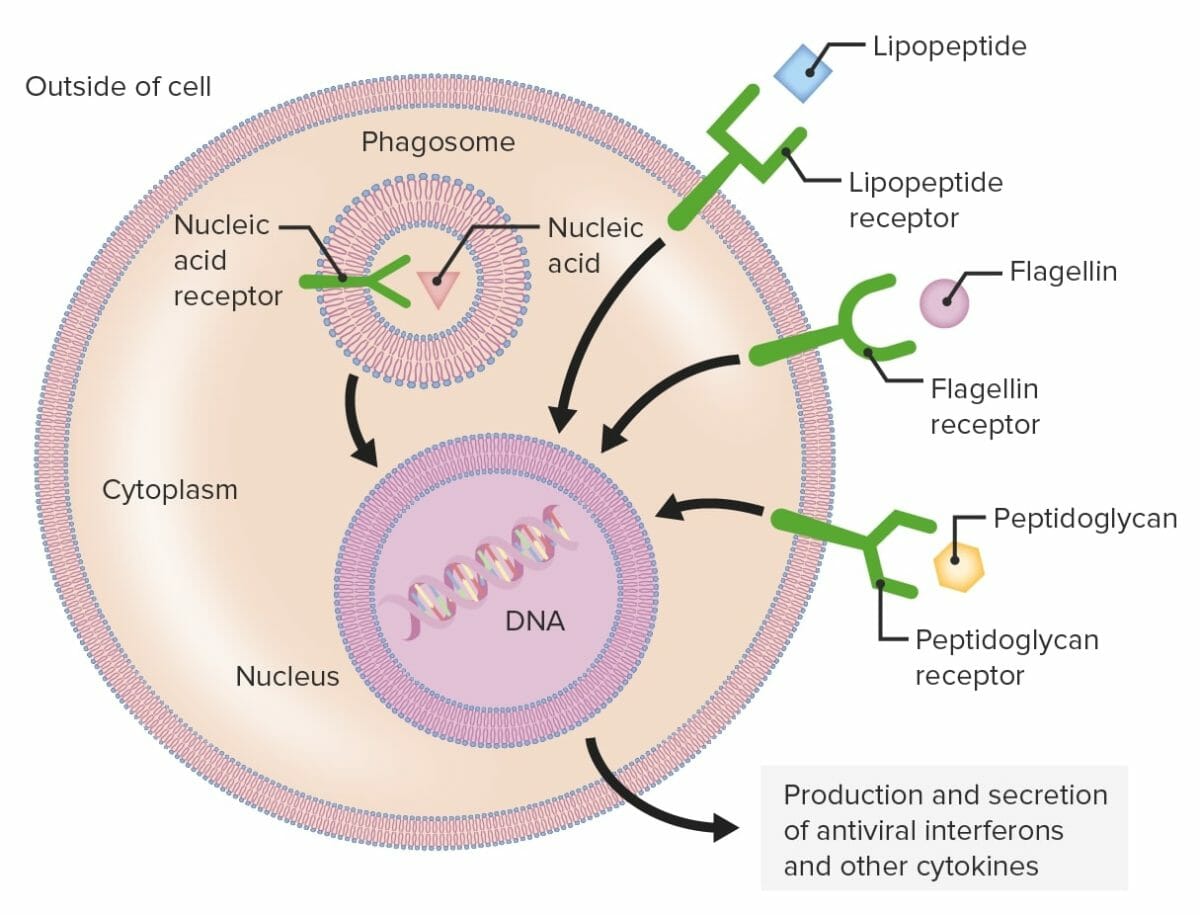
Pattern recognition receptors (PRRs):
Phagocytic cells contain PRRs capable of recognizing various pathogen-associated molecular patterns (PAMPs). Toll-like receptors (TLRs) (shown as green structures), which are a group of PRRs, recognize different microbial components, including lipopeptide, flagellin, or peptidoglycan. The PRRs can be found on the plasma membrane or intracellularly.
When a PRR recognizes a PAMP, a signal is sent to the nucleus which activates genes involved in phagocytosis, cellular proliferation, enhanced intracellular killing, and the production and secretion of antiviral interferons and proinflammatory cytokines.
Secreted and circulating PRRs include many proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis (e.g., AMP, lectins, collectins).
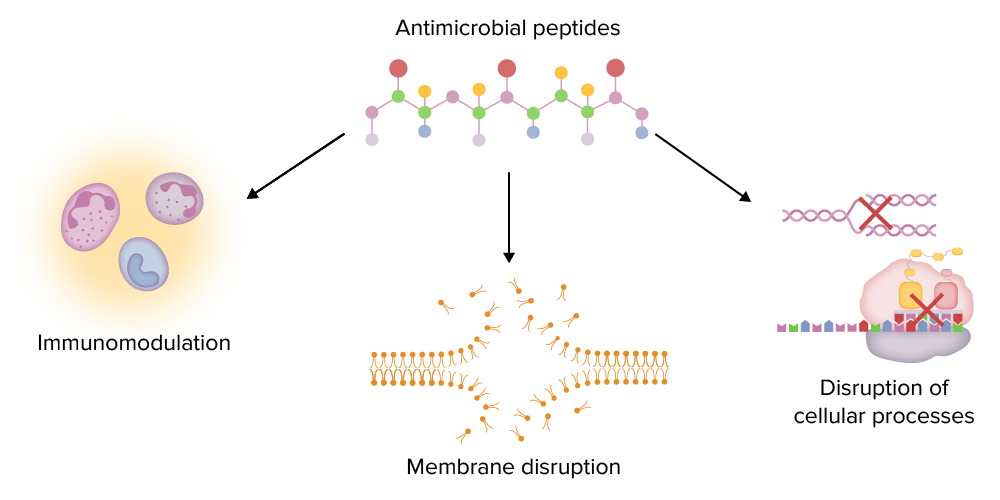
Major functions of antimicrobial peptides (AMPs):
AMPs can modulate the immune system (e.g., stimulating chemotaxis, regulating cytokine production), disrupt microbial membranes, and disrupt important intracellular processes (e.g., DNA and protein synthesis).
Immune responses follow the recognition of pathogen molecules. The complement system is 1 response activated in the cascading fashion to destroy microbes.
Complement activation Complement Activation The sequential activation of serum complement proteins to create the complement membrane attack complex. Factors initiating complement activation include antigen-antibody complexes, microbial antigens, or cell surface polysaccharides. Systemic Lupus Erythematosus is through distinctive pathways (all start with a different initiating molecule), but all produce C3b (the central molecule of the complement cascade Complement cascade The sequential activation of serum complement proteins to create the complement membrane attack complex. Factors initiating complement activation include antigen-antibody complexes, microbial antigens, or cell surface polysaccharides. C3 Deficiency):
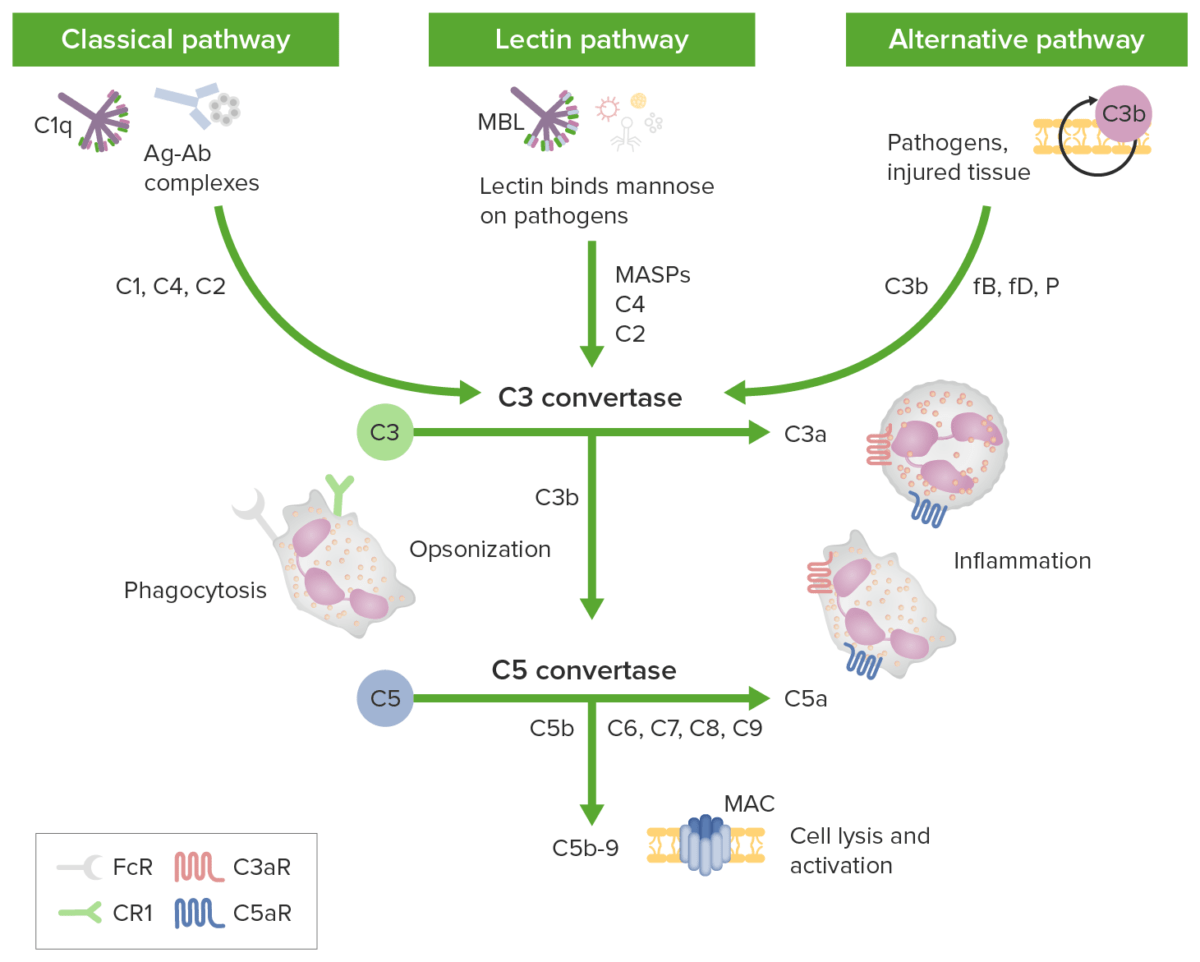
Complement initiation pathways lead to a common terminal pathway:
Green boxes identify initiation pathways; complement components are identified along the arrows. The classical pathway is activated by antigen-antibody complexes (Ag-Ab complexes) recognized by C1q in complex with C1r and C1s. Proteases C1r and C1s cleave C4 and C2 to generate the classical pathway C3 convertase C4b2b. The lectin pathway is triggered by the binding of mannose-binding lectin (MBL) or ficolins to carbohydrates on the target membrane.
The MBL-associated serine proteases (MASPs) then cleave C4 and C2 generating the C3 convertase C4b2b. The alternative pathway is triggered when the low levels of C3b protein directly bind a microbe, foreign material, or damaged tissue. When C3b binds with factor B, C3bB is formed. Factor B is cleaved by factor D to form an alternative pathway C3-convertase (C3bBb). The convertase is stabilized by properdin. C3b opsonizes targets for phagocytosis and B-cell activation.
All 3 initiation pathways converge on C3 with distinct C3 convertases cleaving C3 to generate anaphylatoxin C3a and more C3b to form the C5 convertases (C4b2a3b and C3bBb3b). C5 convertase then cleaves C5 into C5a and C5b. The anaphylatoxins C3a, C4a, and C5a can attract/activate inflammatory cells and contract smooth muscle through receptors C3aR and C5aR. The membrane attack complex (MAC) forms when C5b binds C6, C7, C8, and multiple copies of C9. Membrane attack complex pores can cause cell death by osmotic flux.
Ultimately, the complement pathways aim to eliminate microbes and cellular debris/apoptotic cells:
Cytokines Cytokines Non-antibody proteins secreted by inflammatory leukocytes and some non-leukocytic cells, that act as intercellular mediators. They differ from classical hormones in that they are produced by a number of tissue or cell types rather than by specialized glands. They generally act locally in a paracrine or autocrine rather than endocrine manner. Adaptive Immune Response are soluble proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis released by different cells, which play overlapping roles in innate and adaptive immunity like the complement system.
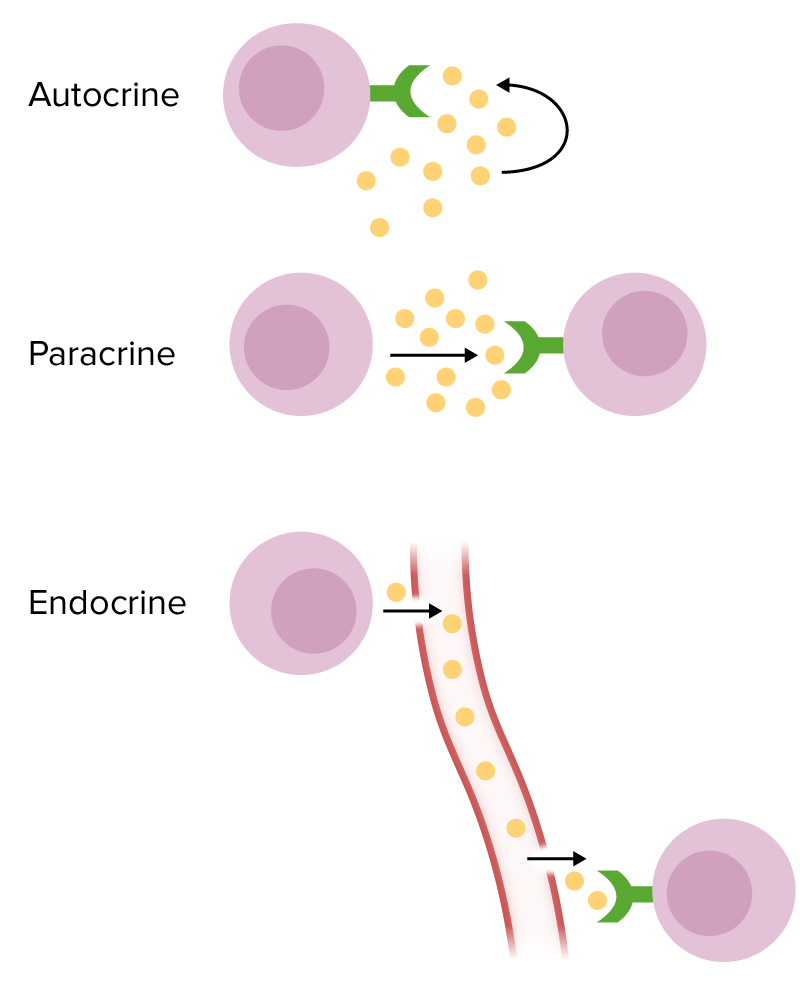
Forms of chemical signaling:
Autocrine: The target cell is the same cell that secretes cytokines.
Paracrine: The target cell for cytokines is a nearby cell.
Endocrine: Cytokines are secreted into the circulation in order to reach a distant target cell.
| Cytokines Cytokines Non-antibody proteins secreted by inflammatory leukocytes and some non-leukocytic cells, that act as intercellular mediators. They differ from classical hormones in that they are produced by a number of tissue or cell types rather than by specialized glands. They generally act locally in a paracrine or autocrine rather than endocrine manner. Adaptive Immune Response | Source | Function and activity |
|---|---|---|
| IL-1 | Monocytes Monocytes Large, phagocytic mononuclear leukocytes produced in the vertebrate bone marrow and released into the blood; contain a large, oval or somewhat indented nucleus surrounded by voluminous cytoplasm and numerous organelles. Innate Immunity: Phagocytes and Antigen Presentation, macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, B cells B cells Lymphoid cells concerned with humoral immunity. They are short-lived cells resembling bursa-derived lymphocytes of birds in their production of immunoglobulin upon appropriate stimulation. B cells: Types and Functions, fibroblasts Fibroblasts Connective tissue cells which secrete an extracellular matrix rich in collagen and other macromolecules. Sarcoidosis, most epithelial cells |
|
| IL-2 | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions |
|
| IL-3 | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, NK cells NK cells A specialized subset of T-lymphocytes that exhibit features of innate immunity similar to that of natural killer cells. They are reactive to glycolipids presented in the context of the major histocompatibility complex (MHC) class I-like molecule, CD1D antigen. Lymphocytes: Histology, mast cells Mast cells Granulated cells that are found in almost all tissues, most abundantly in the skin and the gastrointestinal tract. Like the basophils, mast cells contain large amounts of histamine and heparin. Unlike basophils, mast cells normally remain in the tissues and do not circulate in the blood. Mast cells, derived from the bone marrow stem cells, are regulated by the stem cell factor. Innate Immunity: Phagocytes and Antigen Presentation | Hematopoiesis Hematopoiesis The development and formation of various types of blood cells. Hematopoiesis can take place in the bone marrow (medullary) or outside the bone marrow (extramedullary hematopoiesis). Bone Marrow: Composition and Hematopoiesis progenitor stimulation |
| IL-4 | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, mast cells Mast cells Granulated cells that are found in almost all tissues, most abundantly in the skin and the gastrointestinal tract. Like the basophils, mast cells contain large amounts of histamine and heparin. Unlike basophils, mast cells normally remain in the tissues and do not circulate in the blood. Mast cells, derived from the bone marrow stem cells, are regulated by the stem cell factor. Innate Immunity: Phagocytes and Antigen Presentation, basophils Basophils Granular leukocytes characterized by a relatively pale-staining, lobate nucleus and cytoplasm containing coarse dark-staining granules of variable size and stainable by basic dyes. Innate Immunity: Phagocytes and Antigen Presentation |
|
| IL-5 | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, mast cells Mast cells Granulated cells that are found in almost all tissues, most abundantly in the skin and the gastrointestinal tract. Like the basophils, mast cells contain large amounts of histamine and heparin. Unlike basophils, mast cells normally remain in the tissues and do not circulate in the blood. Mast cells, derived from the bone marrow stem cells, are regulated by the stem cell factor. Innate Immunity: Phagocytes and Antigen Presentation, eosinophils Eosinophils Granular leukocytes with a nucleus that usually has two lobes connected by a slender thread of chromatin, and cytoplasm containing coarse, round granules that are uniform in size and stainable by eosin. Innate Immunity: Phagocytes and Antigen Presentation |
|
| IL-6 | Monocytes Monocytes Large, phagocytic mononuclear leukocytes produced in the vertebrate bone marrow and released into the blood; contain a large, oval or somewhat indented nucleus surrounded by voluminous cytoplasm and numerous organelles. Innate Immunity: Phagocytes and Antigen Presentation, macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, B cells B cells Lymphoid cells concerned with humoral immunity. They are short-lived cells resembling bursa-derived lymphocytes of birds in their production of immunoglobulin upon appropriate stimulation. B cells: Types and Functions, fibroblasts Fibroblasts Connective tissue cells which secrete an extracellular matrix rich in collagen and other macromolecules. Sarcoidosis, most epithelial cells |
|
| IL-7 IL-7 A proinflammatory cytokine produced primarily by T-lymphocytes or their precursors. Several subtypes of interleukin-17 have been identified, each of which is a product of a unique gene. Severe Combined Immunodeficiency (SCID) | Bone marrow Bone marrow The soft tissue filling the cavities of bones. Bone marrow exists in two types, yellow and red. Yellow marrow is found in the large cavities of large bones and consists mostly of fat cells and a few primitive blood cells. Red marrow is a hematopoietic tissue and is the site of production of erythrocytes and granular leukocytes. Bone marrow is made up of a framework of connective tissue containing branching fibers with the frame being filled with marrow cells. Bone Marrow: Composition and Hematopoiesis, thymic epithelial cells | Differentiation of B cells B cells Lymphoid cells concerned with humoral immunity. They are short-lived cells resembling bursa-derived lymphocytes of birds in their production of immunoglobulin upon appropriate stimulation. B cells: Types and Functions, T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, and NK cells NK cells A specialized subset of T-lymphocytes that exhibit features of innate immunity similar to that of natural killer cells. They are reactive to glycolipids presented in the context of the major histocompatibility complex (MHC) class I-like molecule, CD1D antigen. Lymphocytes: Histology |
| IL-8 | Monocytes Monocytes Large, phagocytic mononuclear leukocytes produced in the vertebrate bone marrow and released into the blood; contain a large, oval or somewhat indented nucleus surrounded by voluminous cytoplasm and numerous organelles. Innate Immunity: Phagocytes and Antigen Presentation, macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, neutrophils Neutrophils Granular leukocytes having a nucleus with three to five lobes connected by slender threads of chromatin, and cytoplasm containing fine inconspicuous granules and stainable by neutral dyes. Innate Immunity: Phagocytes and Antigen Presentation, fibroblasts Fibroblasts Connective tissue cells which secrete an extracellular matrix rich in collagen and other macromolecules. Sarcoidosis, endothelial cells, epithelial cells |
|
| IL-9 | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions |
|
| IL-10 | Monocytes Monocytes Large, phagocytic mononuclear leukocytes produced in the vertebrate bone marrow and released into the blood; contain a large, oval or somewhat indented nucleus surrounded by voluminous cytoplasm and numerous organelles. Innate Immunity: Phagocytes and Antigen Presentation, macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, B cells B cells Lymphoid cells concerned with humoral immunity. They are short-lived cells resembling bursa-derived lymphocytes of birds in their production of immunoglobulin upon appropriate stimulation. B cells: Types and Functions, keratinocytes Keratinocytes Epidermal cells which synthesize keratin and undergo characteristic changes as they move upward from the basal layers of the epidermis to the cornified (horny) layer of the skin. Successive stages of differentiation of the keratinocytes forming the epidermal layers are basal cell, spinous or prickle cell, and the granular cell. Skin: Structure and Functions, mast cells Mast cells Granulated cells that are found in almost all tissues, most abundantly in the skin and the gastrointestinal tract. Like the basophils, mast cells contain large amounts of histamine and heparin. Unlike basophils, mast cells normally remain in the tissues and do not circulate in the blood. Mast cells, derived from the bone marrow stem cells, are regulated by the stem cell factor. Innate Immunity: Phagocytes and Antigen Presentation |
|
| IL-11 | Bone marrow Bone marrow The soft tissue filling the cavities of bones. Bone marrow exists in two types, yellow and red. Yellow marrow is found in the large cavities of large bones and consists mostly of fat cells and a few primitive blood cells. Red marrow is a hematopoietic tissue and is the site of production of erythrocytes and granular leukocytes. Bone marrow is made up of a framework of connective tissue containing branching fibers with the frame being filled with marrow cells. Bone Marrow: Composition and Hematopoiesis stromal cells |
|
| IL-12 | Activated macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, dendritic cells Dendritic cells Specialized cells of the hematopoietic system that have branch-like extensions. They are found throughout the lymphatic system, and in non-lymphoid tissues such as skin and the epithelia of the intestinal, respiratory, and reproductive tracts. They trap and process antigens, and present them to T-cells, thereby stimulating cell-mediated immunity. They are different from the non-hematopoietic follicular dendritic cells, which have a similar morphology and immune system function, but with respect to humoral immunity (antibody production). Skin: Structure and Functions, neutrophils Neutrophils Granular leukocytes having a nucleus with three to five lobes connected by slender threads of chromatin, and cytoplasm containing fine inconspicuous granules and stainable by neutral dyes. Innate Immunity: Phagocytes and Antigen Presentation |
|
| IFN-ɣ | T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, NK cells NK cells A specialized subset of T-lymphocytes that exhibit features of innate immunity similar to that of natural killer cells. They are reactive to glycolipids presented in the context of the major histocompatibility complex (MHC) class I-like molecule, CD1D antigen. Lymphocytes: Histology |
|
| TNF-ɑ | Monocytes Monocytes Large, phagocytic mononuclear leukocytes produced in the vertebrate bone marrow and released into the blood; contain a large, oval or somewhat indented nucleus surrounded by voluminous cytoplasm and numerous organelles. Innate Immunity: Phagocytes and Antigen Presentation, macrophages Macrophages The relatively long-lived phagocytic cell of mammalian tissues that are derived from blood monocytes. Main types are peritoneal macrophages; alveolar macrophages; histiocytes; kupffer cells of the liver; and osteoclasts. They may further differentiate within chronic inflammatory lesions to epithelioid cells or may fuse to form foreign body giant cells or langhans giant cells. Innate Immunity: Phagocytes and Antigen Presentation, mast cells Mast cells Granulated cells that are found in almost all tissues, most abundantly in the skin and the gastrointestinal tract. Like the basophils, mast cells contain large amounts of histamine and heparin. Unlike basophils, mast cells normally remain in the tissues and do not circulate in the blood. Mast cells, derived from the bone marrow stem cells, are regulated by the stem cell factor. Innate Immunity: Phagocytes and Antigen Presentation, basophils Basophils Granular leukocytes characterized by a relatively pale-staining, lobate nucleus and cytoplasm containing coarse dark-staining granules of variable size and stainable by basic dyes. Innate Immunity: Phagocytes and Antigen Presentation, eosinophils Eosinophils Granular leukocytes with a nucleus that usually has two lobes connected by a slender thread of chromatin, and cytoplasm containing coarse, round granules that are uniform in size and stainable by eosin. Innate Immunity: Phagocytes and Antigen Presentation, NK cells NK cells A specialized subset of T-lymphocytes that exhibit features of innate immunity similar to that of natural killer cells. They are reactive to glycolipids presented in the context of the major histocompatibility complex (MHC) class I-like molecule, CD1D antigen. Lymphocytes: Histology, B cells B cells Lymphoid cells concerned with humoral immunity. They are short-lived cells resembling bursa-derived lymphocytes of birds in their production of immunoglobulin upon appropriate stimulation. B cells: Types and Functions, T cells T cells Lymphocytes responsible for cell-mediated immunity. Two types have been identified – cytotoxic (t-lymphocytes, cytotoxic) and helper T-lymphocytes (t-lymphocytes, helper-inducer). They are formed when lymphocytes circulate through the thymus gland and differentiate to thymocytes. When exposed to an antigen, they divide rapidly and produce large numbers of new T cells sensitized to that antigen. T cells: Types and Functions, fibroblasts Fibroblasts Connective tissue cells which secrete an extracellular matrix rich in collagen and other macromolecules. Sarcoidosis, thymic epithelial cells |
|
| Transforming growth factor-β | Most cells | Antiinflammatory |
After pathogen recognition and recruitment Recruitment Skeletal Muscle Contraction of immune cells (with coordinated help from complements and cytokines Cytokines Non-antibody proteins secreted by inflammatory leukocytes and some non-leukocytic cells, that act as intercellular mediators. They differ from classical hormones in that they are produced by a number of tissue or cell types rather than by specialized glands. They generally act locally in a paracrine or autocrine rather than endocrine manner. Adaptive Immune Response), strategies are implemented to eliminate the microbes.
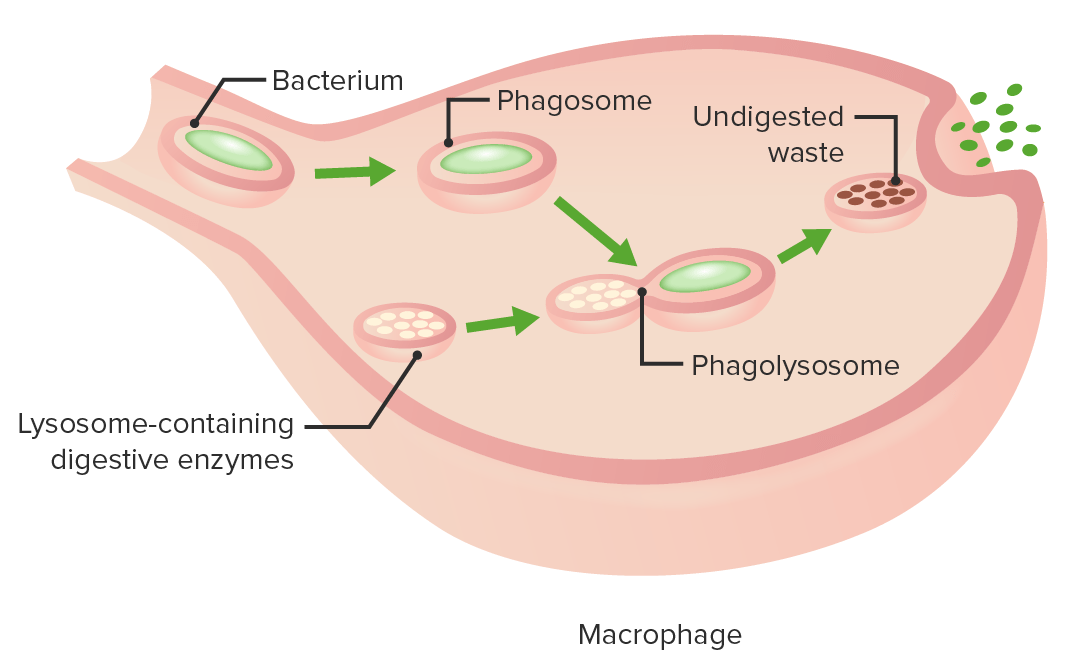
The stages of phagocytosis:
Engulfment of a pathogen, formation of a phagosome, digestion of the pathogenic particle in the phagolysosome, expulsion of undigested materials from the cell
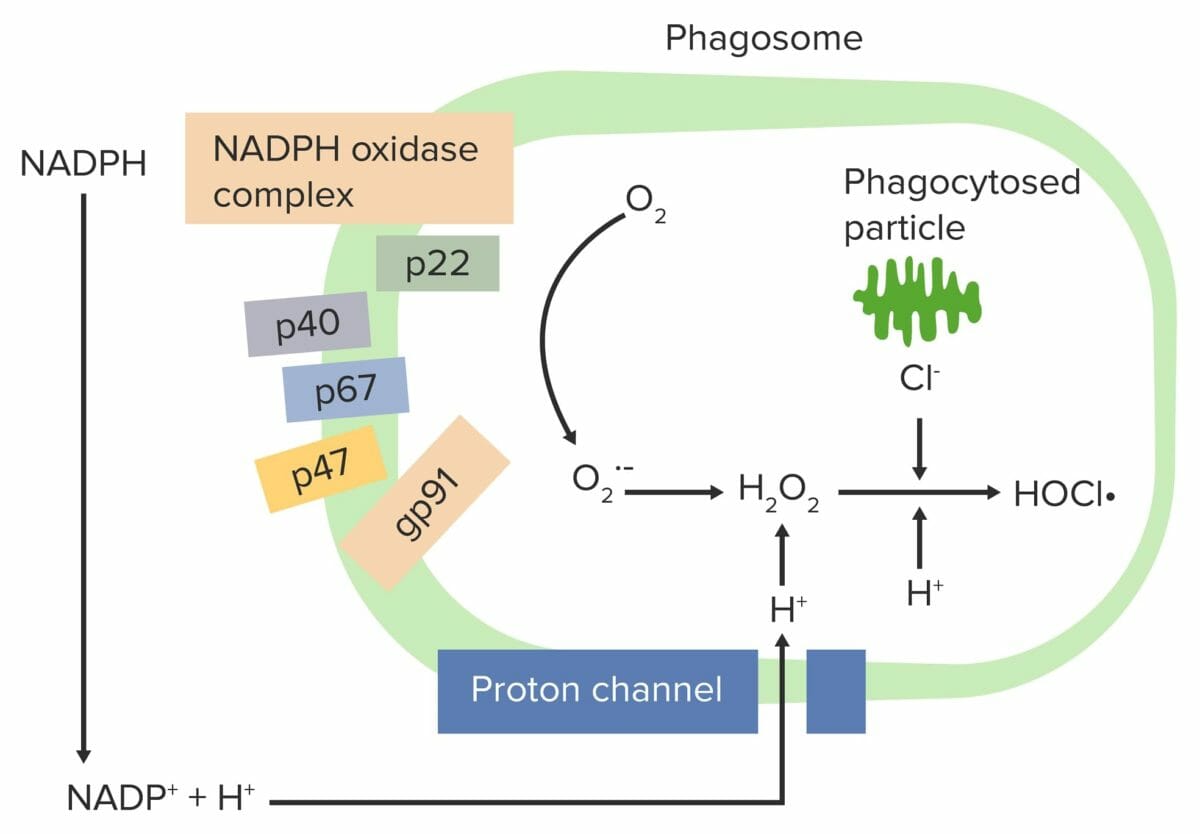
Respiratory burst initiated by the NADPH oxidase complex:
The phagocyte NADPH-oxidase complex is activated, reducing O2 to an oxygen free radical (superoxide anion (O2•–)) and then to H2O2. Neutrophils and monocytes (using myeloperoxidase) combine H2O2 with Cl– to produce hypochlorite (HOCl•), which helps destroy the bacteria.