Viruses are infectious, obligate intracellular parasites composed of a nucleic acid core surrounded by a protein capsid. Viruses can be either naked (non-enveloped) or enveloped. The classification of viruses is complex and based on many factors, including type and structure of the nucleoid and capsid, the presence of an envelope, the replication cycle Cycle The type of signal that ends the inspiratory phase delivered by the ventilator Invasive Mechanical Ventilation, and the host range. The replication cycle Cycle The type of signal that ends the inspiratory phase delivered by the ventilator Invasive Mechanical Ventilation differs between viruses that infect bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology (bacteriophages) and viruses that infect eukaryotic Eukaryotic Eukaryotes can be single-celled or multicellular organisms and include plants, animals, fungi, and protozoa. Eukaryotic cells contain a well-organized nucleus contained by a membrane, along with other membrane-bound organelles. Cell Types: Eukaryotic versus Prokaryotic cells. Bacteriophages have either a lytic or lysogenic replication cycle Cycle The type of signal that ends the inspiratory phase delivered by the ventilator Invasive Mechanical Ventilation, while eukaryotic Eukaryotic Eukaryotes can be single-celled or multicellular organisms and include plants, animals, fungi, and protozoa. Eukaryotic cells contain a well-organized nucleus contained by a membrane, along with other membrane-bound organelles. Cell Types: Eukaryotic versus Prokaryotic viruses have a defined 6-step replication process.
Last updated: Sep 8, 2022
Viruses are infectious, obligate intracellular parasites, composed of a nucleic acid core ( deoxyribonucleic acid Deoxyribonucleic acid A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure ( DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure) or ribonucleic acid Ribonucleic acid A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure ( RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure)) surrounded by a protein capsid; at times, viruses are also surrounded by an envelope derived from host cell membranes.
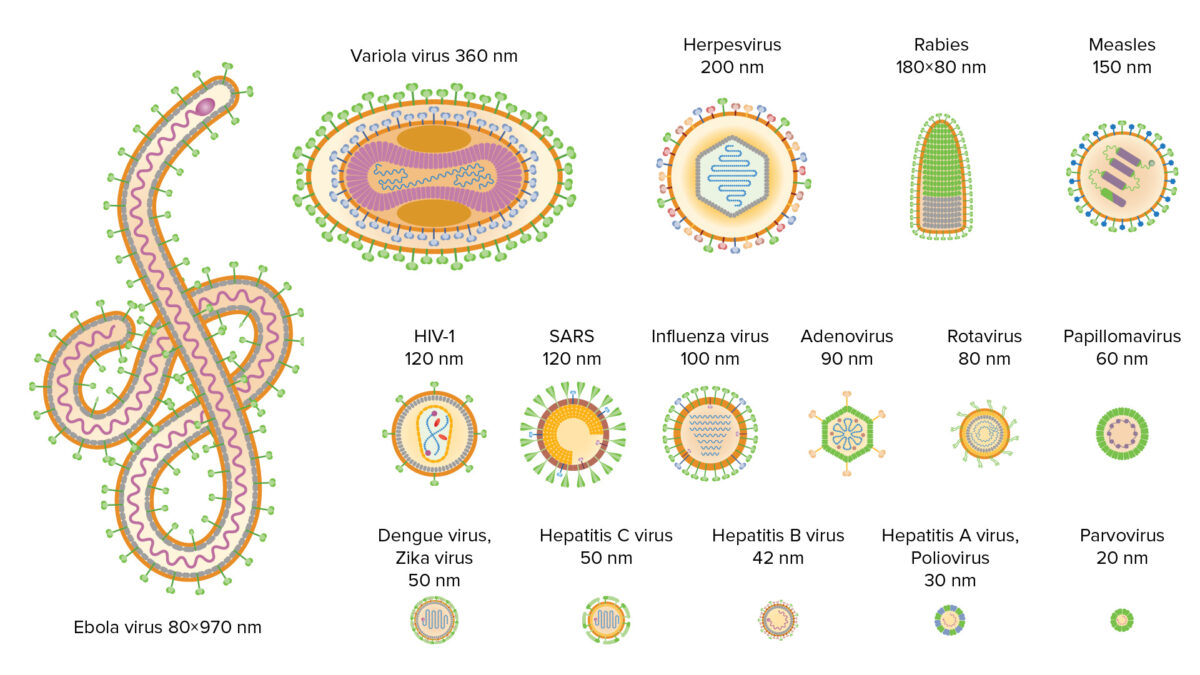
Variety of viruses in existence, including size and morphology
Image by Lecturio. License: CC BY-NC-SA 4.0Basic structure consists of:
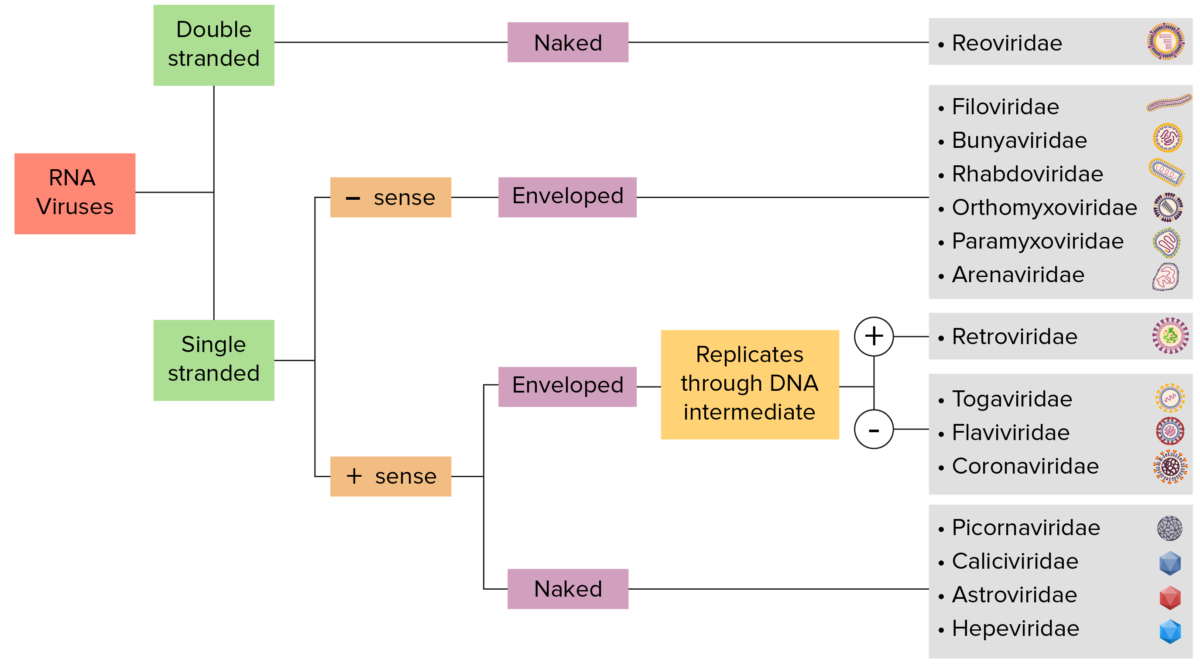
RNA virus identification:
Viruses can be classified in many ways. Most viruses, however, will have a genome formed by either DNA or RNA. RNA genome viruses can be further characterized by either a single- or double-stranded RNA. “Enveloped” viruses are covered by a thin coat of cell membrane (usually taken from the host cell). If the coat is absent, the viruses are called “naked” viruses. Viruses with single-stranded genomes are “positive-sense” viruses if the genome is directly employed as messenger RNA (mRNA), which is translated into proteins. “Negative-sense,” single-stranded viruses employ RNA dependent RNA polymerase, a viral enzyme, to transcribe their genome into messenger RNA.
Multiple categorization Categorization Types of Variables schemes exist for viruses, based on physical characteristics and replication strategies.
| Classification by: | Types |
|---|---|
| Type and structure of nucleoid |
|
| Structure of capsid |
|
| Presence of envelope |
|
| Replication cycle Cycle The type of signal that ends the inspiratory phase delivered by the ventilator Invasive Mechanical Ventilation (for bacteriophages) |
|
| Other |
|
It is important to differentiate between the replication of bacteriophages (viruses that infect bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology) and eukaryotic Eukaryotic Eukaryotes can be single-celled or multicellular organisms and include plants, animals, fungi, and protozoa. Eukaryotic cells contain a well-organized nucleus contained by a membrane, along with other membrane-bound organelles. Cell Types: Eukaryotic versus Prokaryotic viruses (viruses that infect eukaryotic Eukaryotic Eukaryotes can be single-celled or multicellular organisms and include plants, animals, fungi, and protozoa. Eukaryotic cells contain a well-organized nucleus contained by a membrane, along with other membrane-bound organelles. Cell Types: Eukaryotic versus Prokaryotic cells).
There are 2 pathways of replication once a virus is within the bacterial host cell:
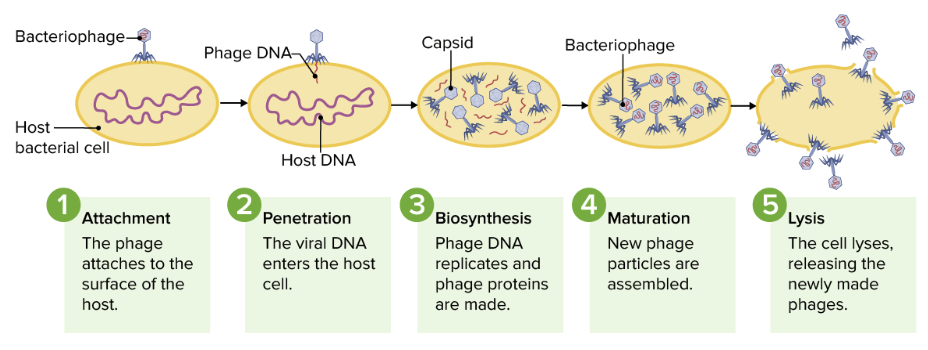
Depiction of the lytic cycle
Image by Lecturio.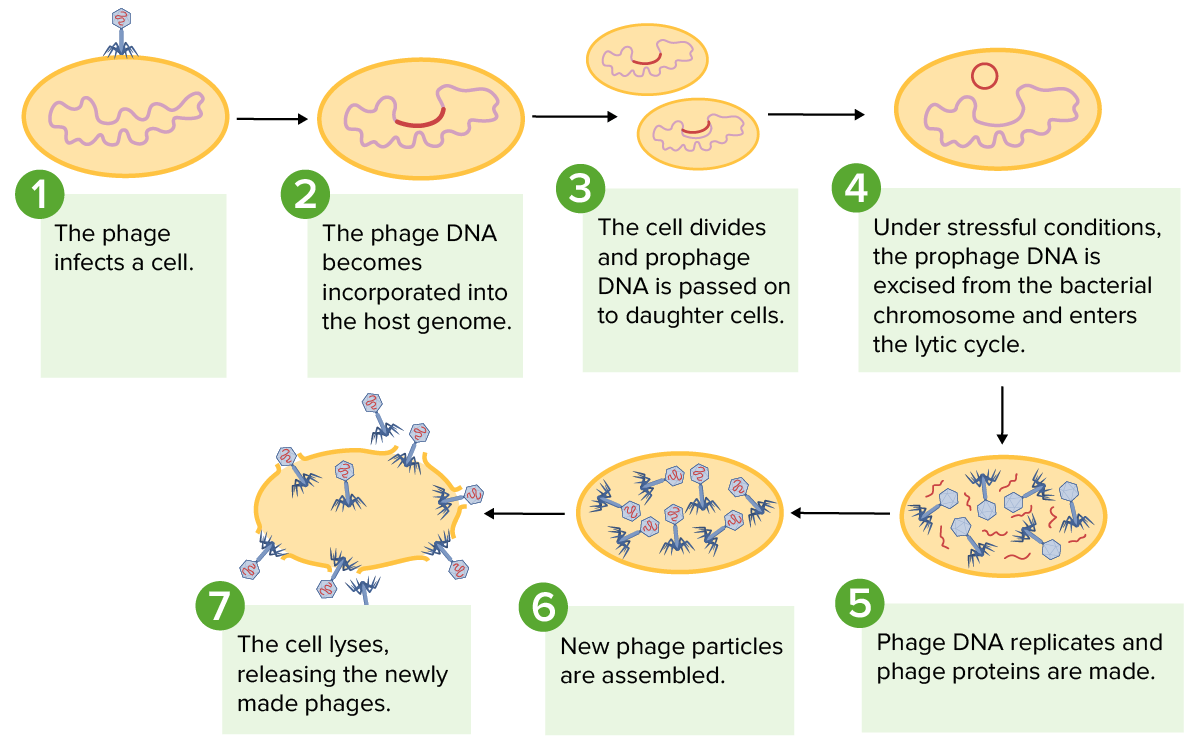
Depiction of the lysogenic and lytic cycles of bacteriophages: During the lysogenic cycle, the bacteriophage DNA is incorporated into the host genome, which can be passed to daughter cells. Stressors to a cell can cause entry into the lytic cycle, where the phage DNA replicates, forms new phage particles, and causes cell lysis.
Image by Lecturio.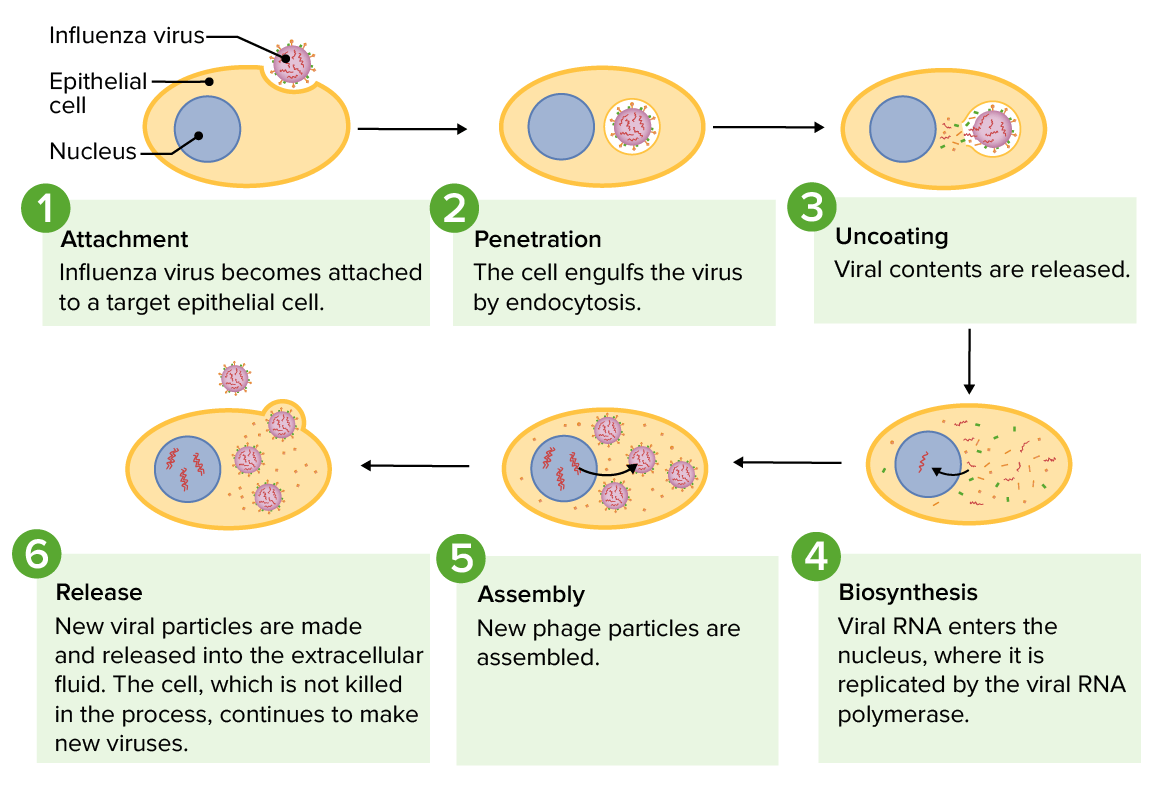
The replication cycle of the influenza virus from infection of a host cell to release of newly formed viruses
Image by Lecturio.Viruses have evolved many processes to increase their genetic diversity:
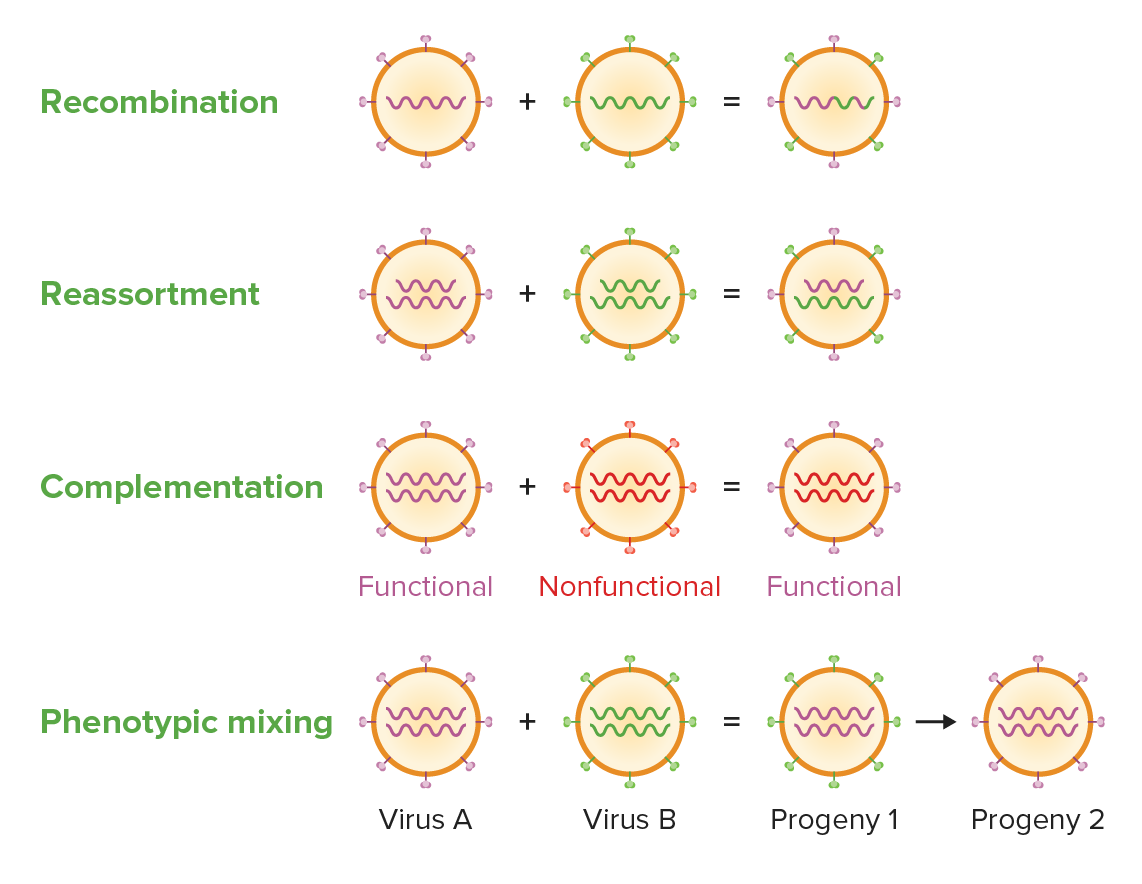
Viruses have evolved diverse strategies for increasing genetic diversity including recombination (the exchange of genes between 2 chromosomes by crossover at homologous regions), reassortment (exchange of segments of chromosomes), complementation (the exchange of genetic material from a functional virus with a nonfunctional virus to convert the latter into functional) and phenotypic mixing (mixing of 2 viral genomes a simultaneously infected host cell giving rise to progeny with blended genomes).
Image by Lecturio. License: CC BY-NC-SA 4.0