Ribonucleic acid (RNA), like deoxyribonucleic acid Deoxyribonucleic acid A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure ( DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure), is a polymer of nucleotides Nucleotides The monomeric units from which DNA or RNA polymers are constructed. They consist of a purine or pyrimidine base, a pentose sugar, and a phosphate group. Nucleic Acids that is essential to cellular protein synthesis Synthesis Polymerase Chain Reaction (PCR). Unlike DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure, RNA is a single-stranded structure containing the sugar moiety ribose Ribose A pentose active in biological systems usually in its d-form. Nucleic Acids (instead of deoxyribose Deoxyribose Nucleic Acids) and the base uracil Uracil One of four nucleotide bases in the nucleic acid RNA. Nucleic Acids (instead of thymine Thymine One of four constituent bases of DNA. Nucleic Acids). While DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure stores the genetic information, RNA generally carries out the instructions encoded in the DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure but RNA also executes diverse non-coding functions. There are 3 major types of RNA that perform different but collaborative roles in protein synthesis Synthesis Polymerase Chain Reaction (PCR): messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA). During transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription, RNA is synthesized from DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure through a series of steps catalyzed by the enzyme RNA polymerase. The mRNA formed will serve as an amino acid Amino acid Amino acids (AAs) are composed of a central carbon atom attached to a carboxyl group, an amino group, a hydrogen atom, and a side chain (R group). Basics of Amino Acids template for protein synthesis Synthesis Polymerase Chain Reaction (PCR). Translation Translation Translation is the process of synthesizing a protein from a messenger RNA (mRNA) transcript. This process is divided into three primary stages: initiation, elongation, and termination. Translation is catalyzed by structures known as ribosomes, which are large complexes of proteins and ribosomal RNA (rRNA). Stages and Regulation of Translation proceeds with the tRNA transporting the corresponding amino acid Amino acid Amino acids (AAs) are composed of a central carbon atom attached to a carboxyl group, an amino group, a hydrogen atom, and a side chain (R group). Basics of Amino Acids based on the deciphered nucleotide sequence ( codon Codon A set of three nucleotides in a protein coding sequence that specifies individual amino acids or a termination signal. Most codons are universal, but some organisms do not produce the transfer RNAs complementary to all codons. These codons are referred to as unassigned codons. Basic Terms of Genetics) in the mRNA. The ribosomes Ribosomes Multicomponent ribonucleoprotein structures found in the cytoplasm of all cells, and in mitochondria, and plastids. They function in protein biosynthesis via genetic translation. The Cell: Organelles, which are composed of rRNA, then facilitate the assembly of amino acids Amino acids Organic compounds that generally contain an amino (-NH2) and a carboxyl (-COOH) group. Twenty alpha-amino acids are the subunits which are polymerized to form proteins. Basics of Amino Acids into a polypeptide. These components work together to convert the mRNA template obtained from DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure into the desired protein.
Last updated: May 18, 2025
Ribonucleic acid (RNA) is a single-stranded polymer of nucleotides Nucleotides The monomeric units from which DNA or RNA polymers are constructed. They consist of a purine or pyrimidine base, a pentose sugar, and a phosphate group. Nucleic Acids that are linked through 3’–5’ phosphodiester bonds.
Nucleotides Nucleotides The monomeric units from which DNA or RNA polymers are constructed. They consist of a purine or pyrimidine base, a pentose sugar, and a phosphate group. Nucleic Acids are the basic unit of any nucleic acid. The nucleotides Nucleotides The monomeric units from which DNA or RNA polymers are constructed. They consist of a purine or pyrimidine base, a pentose sugar, and a phosphate group. Nucleic Acids in RNA are formed by the following parts:
Nucleic acid components:
| RNA | DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure | |
|---|---|---|
| Sugar moiety | Ribose Ribose A pentose active in biological systems usually in its d-form. Nucleic Acids | Deoxyribose Deoxyribose Nucleic Acids |
| Nitrogenous bases Nitrogenous bases Nucleic Acids |
|
|
| Basic structure | Single stranded | Double stranded |
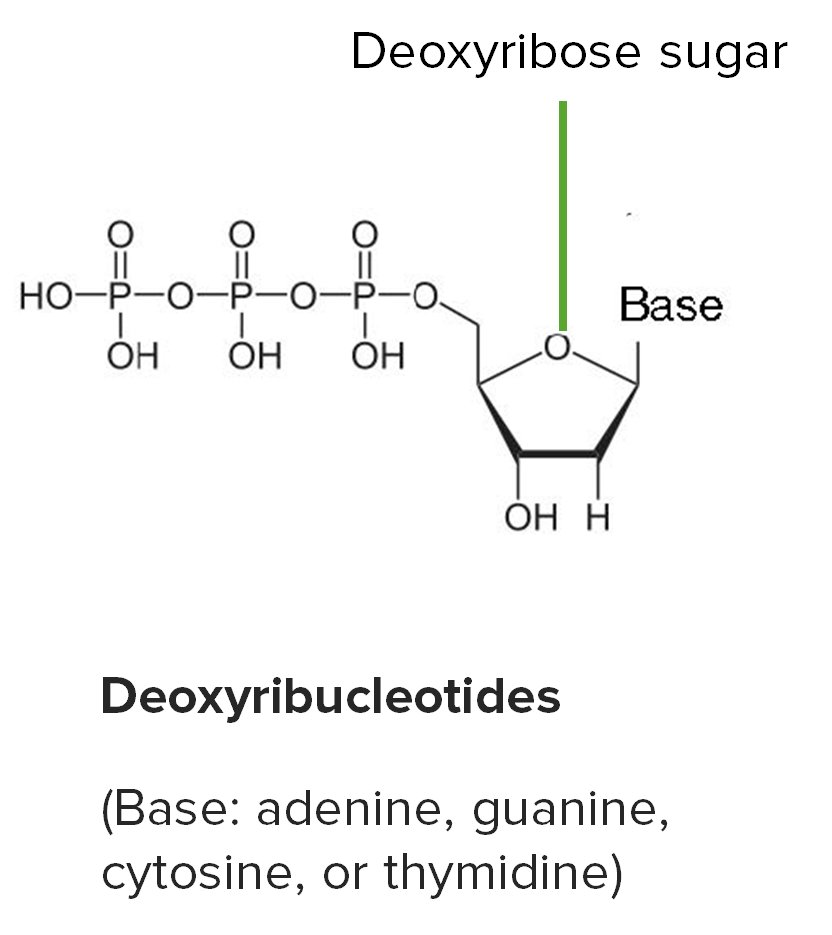
Basic structure of ribonucleotides: carbon 2’ is bonded to hydroxyl (OH) in RNA.
Image by Lecturio.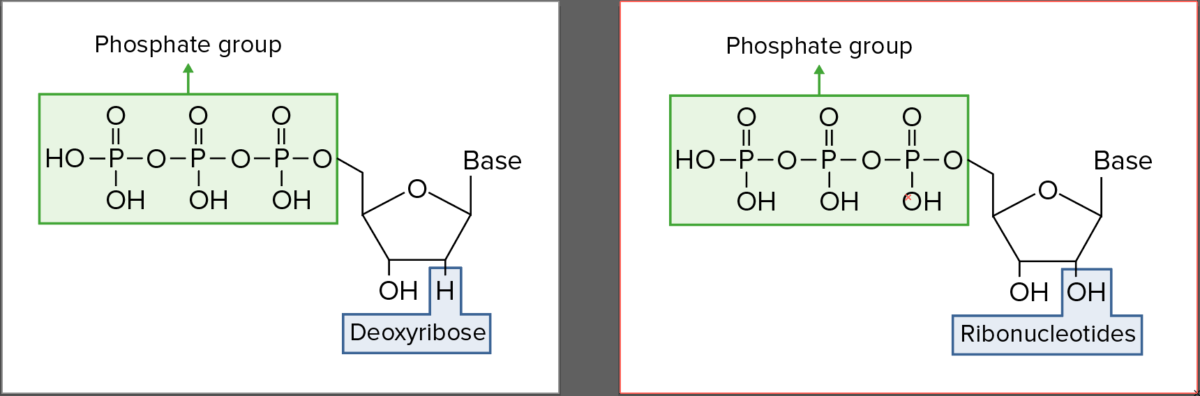
Structures of each ribonucleotide
Deoxyribonucleotides:
1. Phosphate group
2. Deoxyribose (Note: Carbon 2′ is bonded to the H.)
3. Nitrogenous base (adenine, thymine, guanine, cytosine)
Ribonucleotides:
1. Phosphate group
2. Ribose (Note: Carbon 2′ is bonded to OH.)
3. Nitrogenous base (adenine, thymine, guanine, cytosine)
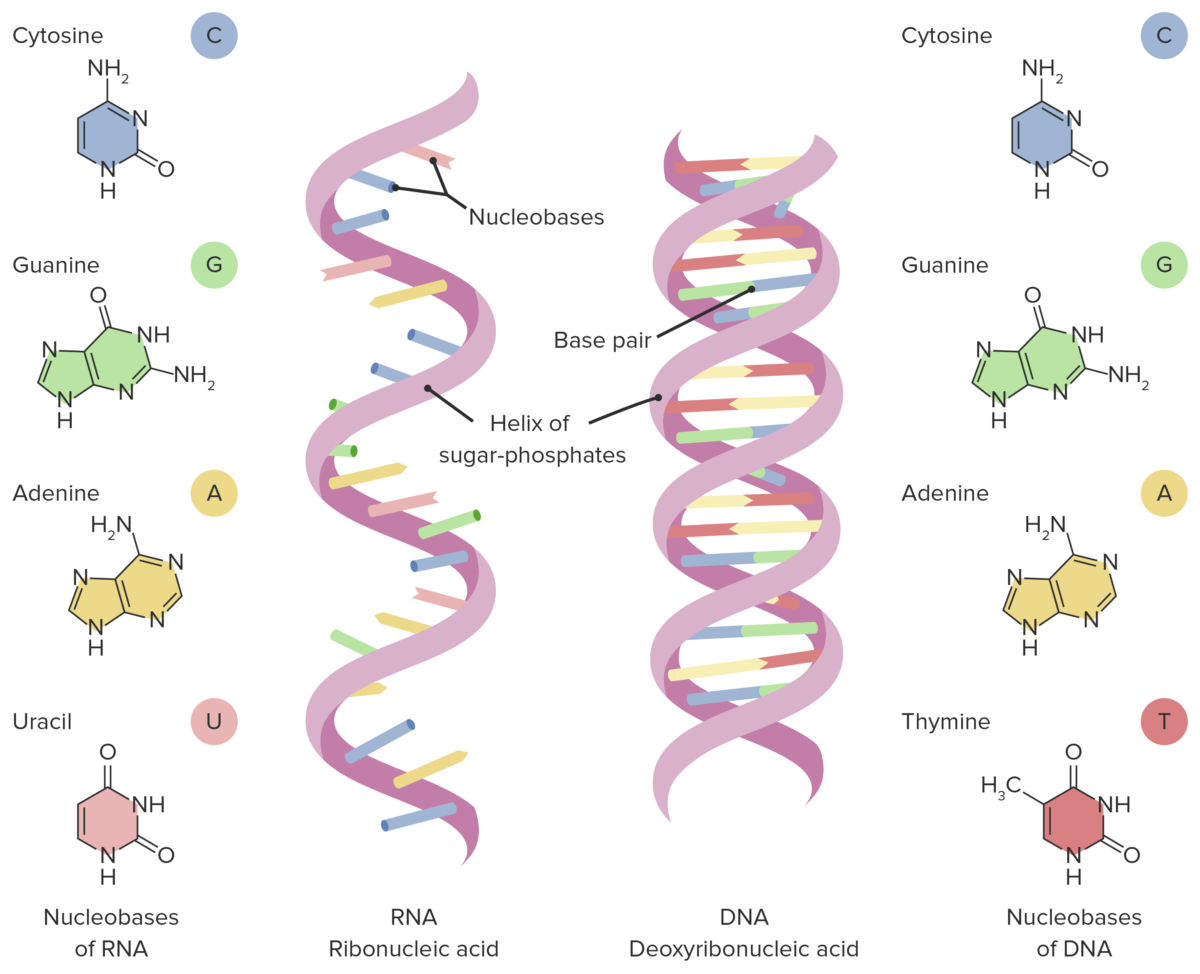
Differences between the structure of DNA and RNA. Note: RNA is a single-stranded structure and has uracil (instead of thymine) as the base.
Image by Lecturio.Coding versus non-coding RNA
Mnemonic
To remember mRNA start codons, use the mnemonic AUG:
To remember mRNA stop codons, use the mnemonics UGA, UAA, and UAG:
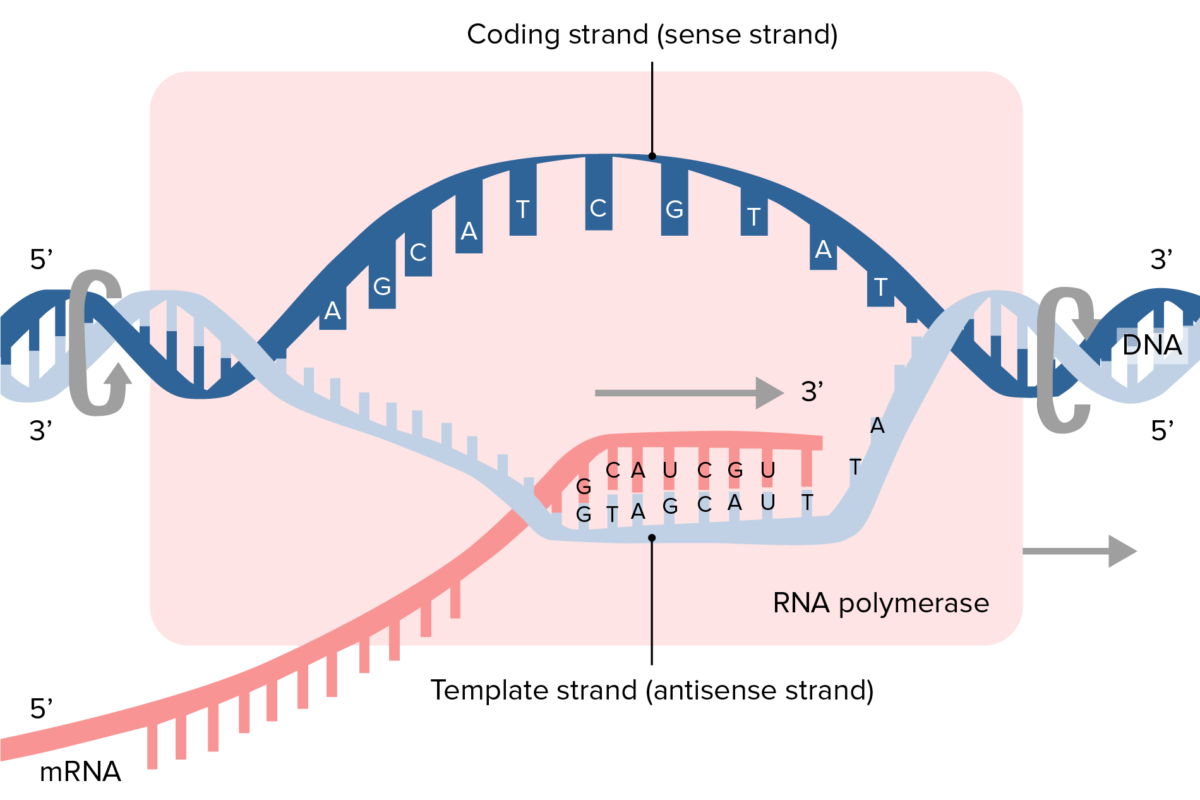
Illustration of transcription process and synthesis of mRNA
Image by Lecturio.Messenger RNA (mRNA) serves as an amino acid Amino acid Amino acids (AAs) are composed of a central carbon atom attached to a carboxyl group, an amino group, a hydrogen atom, and a side chain (R group). Basics of Amino Acids template for protein synthesis Synthesis Polymerase Chain Reaction (PCR).
Prokaryotic Prokaryotic Prokaryotes are unicellular organisms that include 2 of the 3 domains of life: bacteria and archaea. Prokaryotic cells consist of a single cytoplasm-filled compartment enclosed by a cell membrane and cell wall. Cell Types: Eukaryotic versus Prokaryotic mRNA
Eukaryotic Eukaryotic Eukaryotes can be single-celled or multicellular organisms and include plants, animals, fungi, and protozoa. Eukaryotic cells contain a well-organized nucleus contained by a membrane, along with other membrane-bound organelles. Cell Types: Eukaryotic versus Prokaryotic mRNA
There is a long-held belief that prokaryotes lack the 5’ cap. Recently, however, some bacteria Bacteria Bacteria are prokaryotic single-celled microorganisms that are metabolically active and divide by binary fission. Some of these organisms play a significant role in the pathogenesis of diseases. Bacteriology have been found to have a 5’ nicotinamide adenine dinucleotide Nicotinamide adenine dinucleotide A coenzyme composed of ribosylnicotinamide 5′-diphosphate coupled to adenosine 5′-phosphate by pyrophosphate linkage. It is found widely in nature and is involved in numerous enzymatic reactions in which it serves as an electron carrier by being alternately oxidized (NAD+) and reduced (NADH). Pentose Phosphate Pathway ( NAD NAD+ A coenzyme composed of ribosylnicotinamide 5′-diphosphate coupled to adenosine 5′-phosphate by pyrophosphate linkage. It is found widely in nature and is involved in numerous enzymatic reactions in which it serves as an electron carrier by being alternately oxidized (NAD+) and reduced (NADH). Pentose Phosphate Pathway) cap:
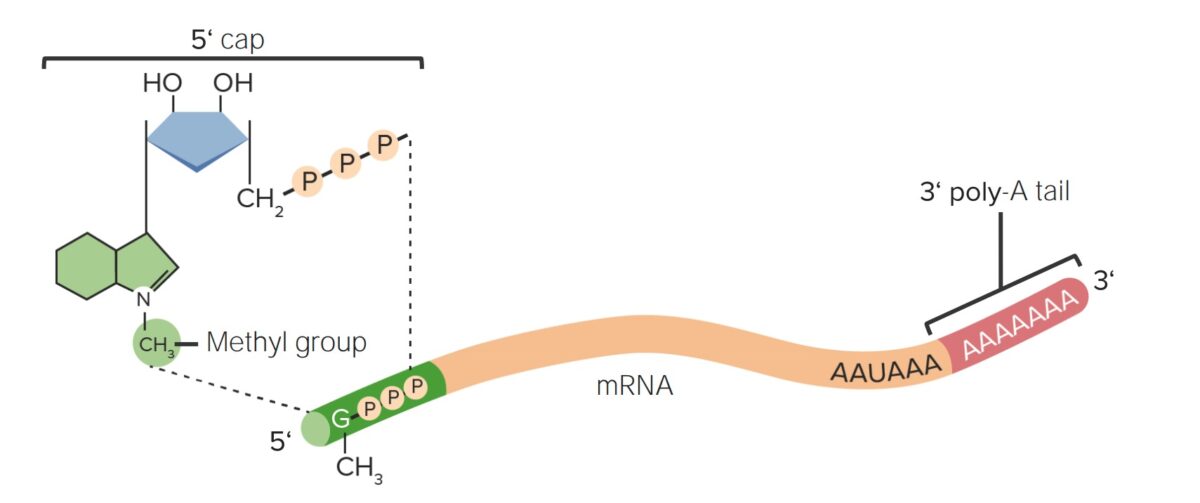
Overview of segments of messenger RNA (mRNA): including the 5’ cap and 3’ poly-A tail
Image by Lecturio.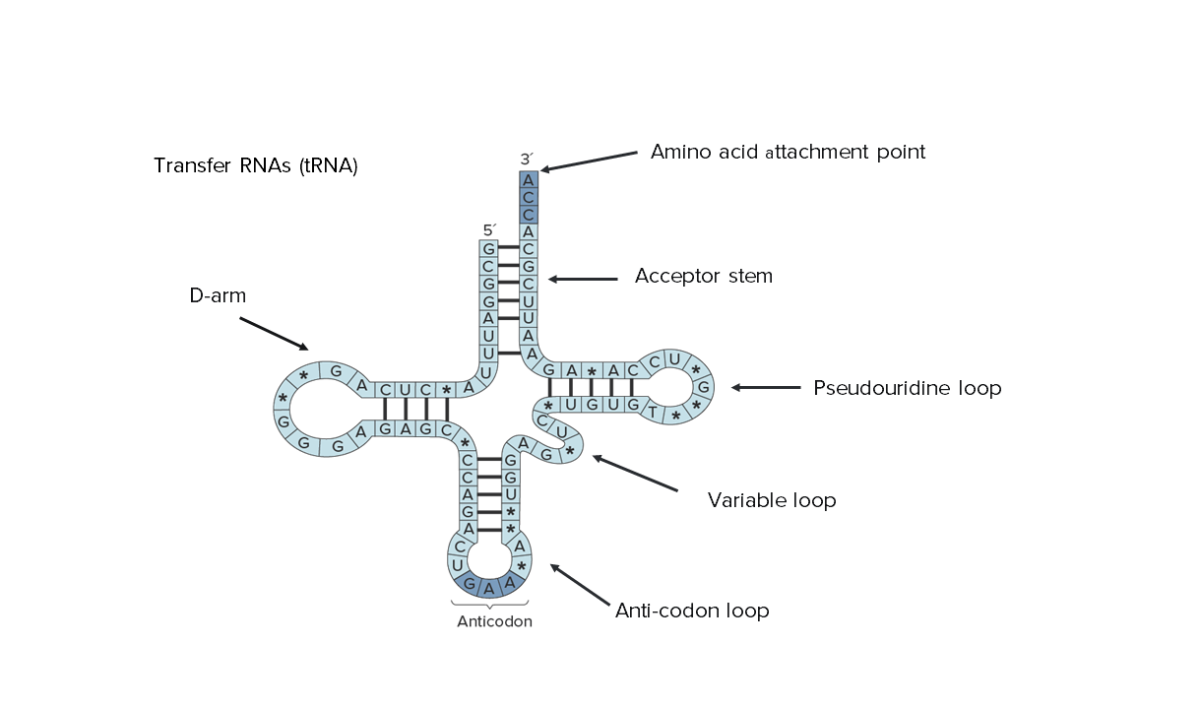
Secondary structure of transfer RNA (tRNA): Notice that its entire sequence can be seen, indicating the reduced size.
Image by Lecturio.Transfer RNA transports amino acids Amino acids Organic compounds that generally contain an amino (-NH2) and a carboxyl (-COOH) group. Twenty alpha-amino acids are the subunits which are polymerized to form proteins. Basics of Amino Acids to ribosomes Ribosomes Multicomponent ribonucleoprotein structures found in the cytoplasm of all cells, and in mitochondria, and plastids. They function in protein biosynthesis via genetic translation. The Cell: Organelles for assembly into proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis. This process is carried out by these 2 main actions:
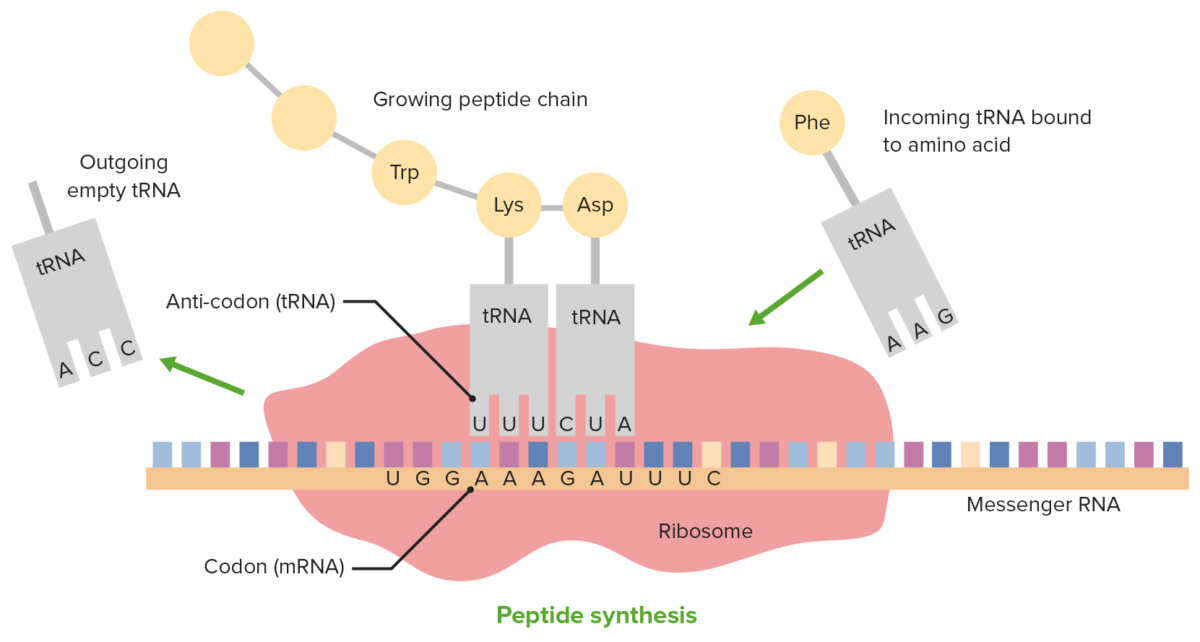
Translation and the role of tRNA
Image by Lecturio.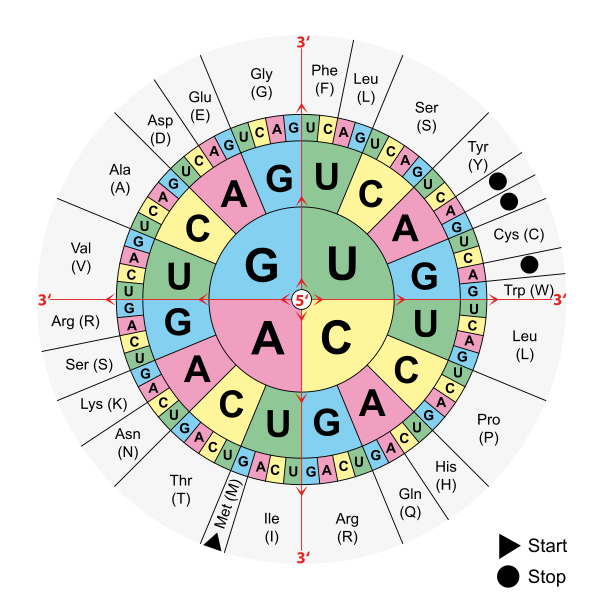
The degeneracy of the genetic code as shown by this codon wheel. Note that many amino acids are encoded by more than one combination of bases.
Image: “Aminoacids table” by Mouagip. License: Public DomainRibosomes consist of 2 subunits of unequally sized rRNA, and the sizes are measured in terms of an “S” value (Svedberg or sedimentation unit):
| Subunits | rRNA | Function |
|---|---|---|
| 50S | 5S | Transmit and coordinate functional centers of ribosome |
| 23S | Peptidyl transferase Peptidyl Transferase Chloramphenicol: peptide bond Peptide bond A peptide bond is formed when the alpha carboxyl group of one amino acid reacts with the alpha-amino group of another amino acid. Proteins and Peptides formation | |
| 30S | 16S | Bind BIND Hyperbilirubinemia of the Newborn initiation codon Codon A set of three nucleotides in a protein coding sequence that specifies individual amino acids or a termination signal. Most codons are universal, but some organisms do not produce the transfer RNAs complementary to all codons. These codons are referred to as unassigned codons. Basic Terms of Genetics; ribosomal scaffold |
| Subunits | rRNA | Function |
|---|---|---|
| 60S | 5S | Structural support |
| 5.8S | Translation Translation Translation is the process of synthesizing a protein from a messenger RNA (mRNA) transcript. This process is divided into three primary stages: initiation, elongation, and termination. Translation is catalyzed by structures known as ribosomes, which are large complexes of proteins and ribosomal RNA (rRNA). Stages and Regulation of Translation | |
| 28S | Peptidyl transferase Peptidyl Transferase Chloramphenicol: peptide bond Peptide bond A peptide bond is formed when the alpha carboxyl group of one amino acid reacts with the alpha-amino group of another amino acid. Proteins and Peptides formation | |
| 40S | 18S | Translation Translation Translation is the process of synthesizing a protein from a messenger RNA (mRNA) transcript. This process is divided into three primary stages: initiation, elongation, and termination. Translation is catalyzed by structures known as ribosomes, which are large complexes of proteins and ribosomal RNA (rRNA). Stages and Regulation of Translation |
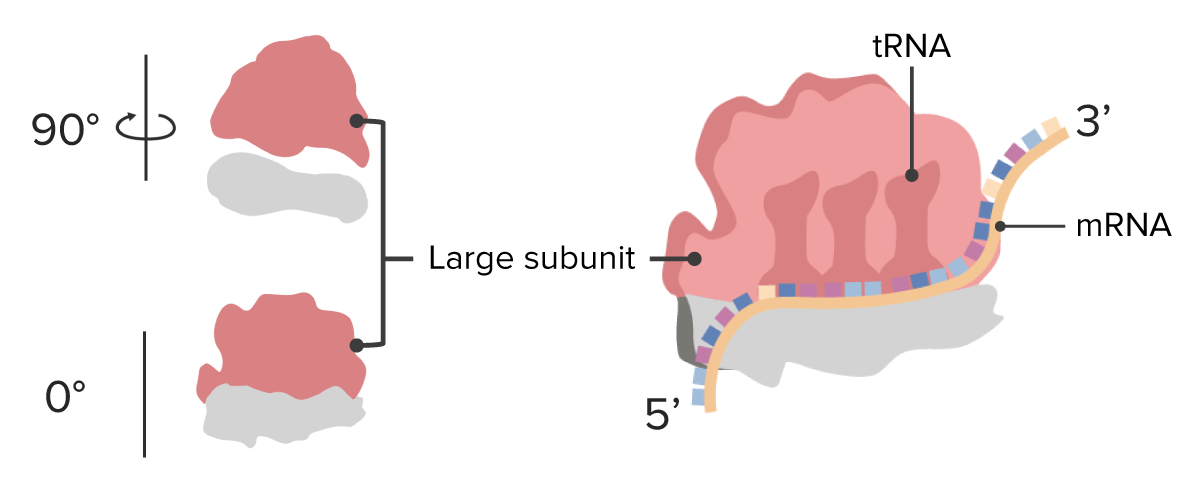
Ribosome: large subunits
Image by Lecturio.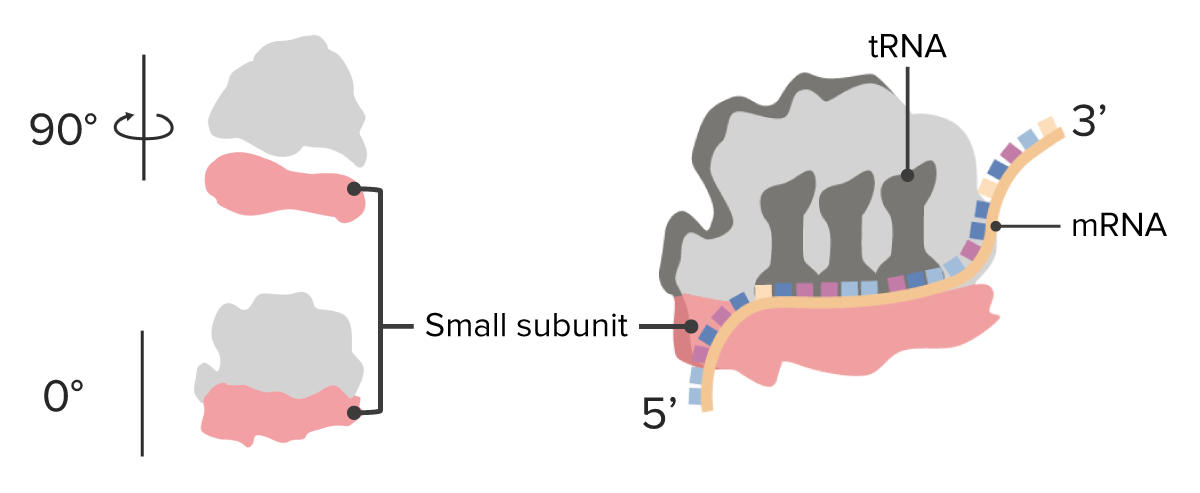
Ribosome: smallsubunits
Image by Lecturio.Small nuclear RNA:
Small nucleolar RNA:
Micro RNA:
Small regulatory RNA:
6sRNA:
Small interfering RNA:
PIWI-interacting RNA:
CRISPR (crRNA):
Ribonuclease Ribonuclease Enzymes that catalyze the hydrolysis of ester bonds within RNA. Interferons P (RNAse P):
Self-splicing introns Introns Sequences of DNA in the genes that are located between the exons. They are transcribed along with the exons but are removed from the primary gene transcript by RNA splicing to leave mature RNA. Some introns code for separate genes. Post-transcriptional Modifications (RNA Processing):
Viroid: