Post-transcriptional modifications Post-transcriptional Modifications Post-transcriptional biological modification of messenger, transfer, or ribosomal RNAs or their precursors. It includes cleavage, methylation, thiolation, isopentenylation, pseudouridine formation, conformational changes, and association with ribosomal protein. Stages of Transcription (PTMs) are processes that facilitate the generation of mature, functional RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure. These rapidly responsive regulatory mechanisms allow different proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis to be produced from one gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics and act as regulators of the phenotype Phenotype The complete genetic complement contained in the DNA of a set of chromosomes in a human. The length of the human genome is about 3 billion base pairs. Basic Terms of Genetics and proliferation rate. These modifications also play a role in some forms of cancer and neurodegenerative diseases. The pre-messenger RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure ( mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3' end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure), called heterogeneous nuclear RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure (hnRNA), is modified by adding a 5’ 7-methylguanosine cap and a 3’ poly-A (polyadenylate) tail for stability and protection. Moreover, hnRNA that contains introns (noncoding sequences) among the expressed sequences or exons undergo splicing. This process removes introns to produce a mature mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3' end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure carrying the coding sequence for translation Translation Translation is the process of synthesizing a protein from a messenger RNA (mRNA) transcript. This process is divided into three primary stages: initiation, elongation, and termination. Translation is catalyzed by structures known as ribosomes, which are large complexes of proteins and ribosomal RNA (rRNA). Stages and Regulation of Translation. Alternative splicing, on the other hand Hand The hand constitutes the distal part of the upper limb and provides the fine, precise movements needed in activities of daily living. It consists of 5 metacarpal bones and 14 phalanges, as well as numerous muscles innervated by the median and ulnar nerves. Hand: Anatomy, also excludes the introns, but varying combinations of exons are linked, producing different proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis from the original mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3' end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure. In RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure editing, the mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3' end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure sequence is altered and differs from the transcribed DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure template. Transfer RNA Transfer RNA The small RNA molecules, 73-80 nucleotides long, that function during translation to align amino acids at the ribosomes in a sequence determined by the mRNA (messenger RNA). There are about 30 different transfer RNAs. Each recognizes a specific codon set on the mRNA through its own anticodon and as aminoacyl tRNAs, each carries a specific amino acid to the ribosome to add to the elongating peptide chains. RNA Types and Structure and ribosomal RNA Ribosomal RNA The most abundant form of RNA. Together with proteins, it forms the ribosomes, playing a structural role and also a role in ribosomal binding of mRNA and tRNAs. Individual chains are conventionally designated by their sedimentation coefficients. In eukaryotes, four large chains exist, synthesized in the nucleolus and constituting about 50% of the ribosome. RNA Types and Structure start from longer precursor molecules and go through steps that include methylation Methylation Addition of methyl groups. In histo-chemistry methylation is used to esterify carboxyl groups and remove sulfate groups by treating tissue sections with hot methanol in the presence of hydrochloric acid. . Glucocorticoids, trimming, and addition of nucleotides Nucleotides The monomeric units from which DNA or RNA polymers are constructed. They consist of a purine or pyrimidine base, a pentose sugar, and a phosphate group. Nucleic Acids.
Last updated: Apr 25, 2025
Genetic information from DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure is copied into messenger RNA Messenger RNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3′ end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure ( mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3′ end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure).
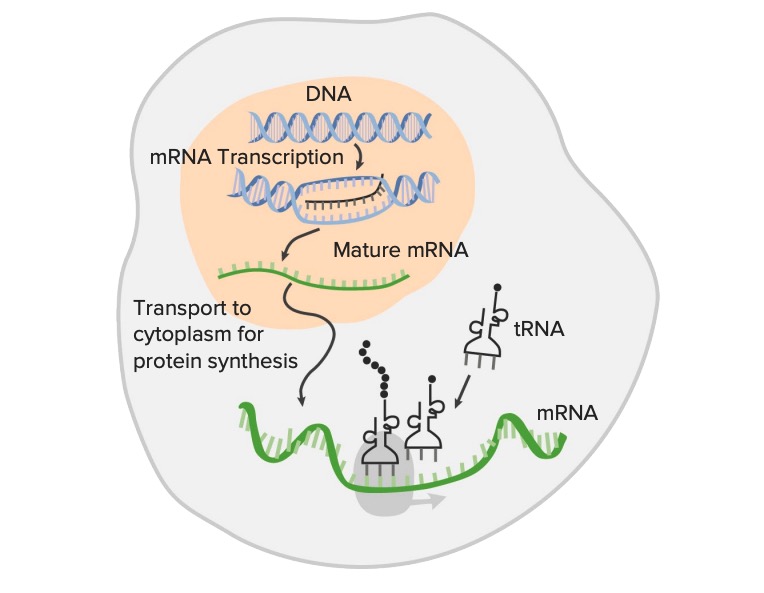
Gene expression from DNA, the genetic sequence, is transcribed into the RNA (transcription):
Transcription of genetic information is the first step in gene expression and is the process through which a coding region of DNA (double-stranded structure) is used as a template for the synthesis of messenger RNA (mRNA). The mature mRNA is translated into amino acids, forming proteins (translation) with the help of ribosomal RNA and transfer RNA (tRNA). This image shows transcription without post-transcriptional modifications of the RNA.
Primary transcripts, or immediate products of transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription, undergo alterations to become biologically functional.
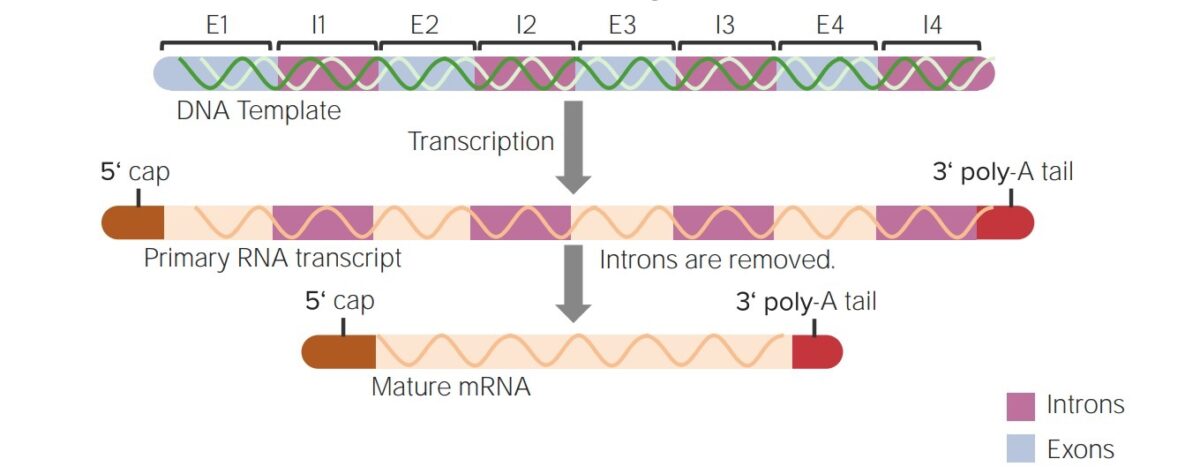
Summary of post-transcriptional modifications of hnRNA into a mature mRNA:
The addition of the 5’ cap and the 3’ poly-A tail and splicing (removal of the intervening sequences or introns)
7-Methylguanosine (methylated guanylyl residue) is added to the 5’ end of hnRNA via:
Functions:
50 to 250 adenylyl residues (AMP) are added to the 3’ end of hnRNA via:
Function:
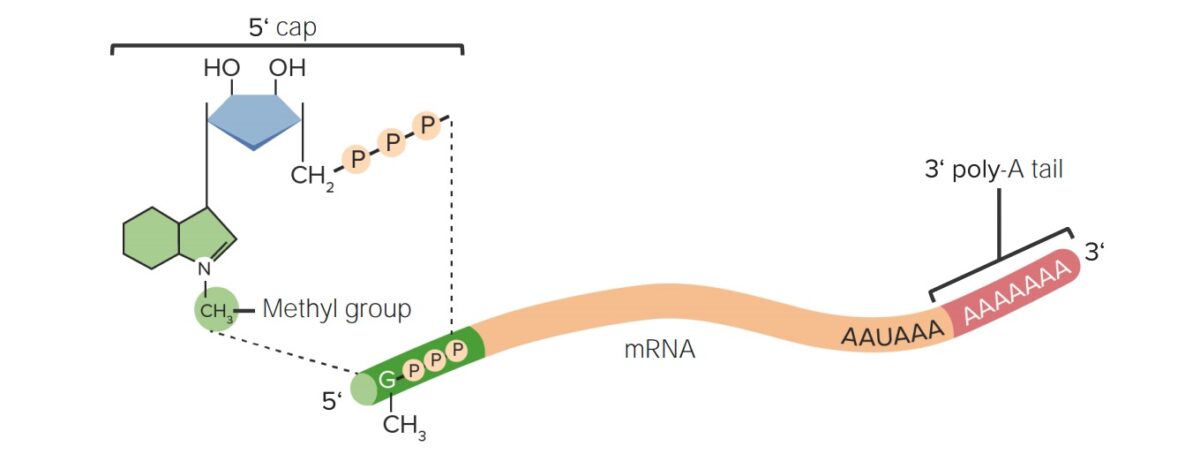
Post-transcriptional modifications of RNA:
The 5’ cap (7-methylguanosine) and 3’ poly-A tail modifications prevent degradation of the mRNA in the cytosol.
Heterogeneous nuclear (pre-mRNA) contains:
Processing:
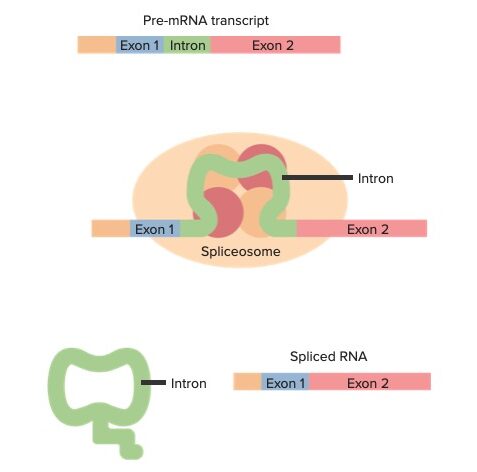
Pre-mRNA exons and introns with an overview of splicing (from top to bottom):
Pre-mRNA transcript contains exons and introns. Interactions of the transcript with small nuclear ribonucleoproteins and other proteins form a spliceosome at certain junctions of the transcript. Cuts are made at the splice sites, and the intron is released. Spliced RNA now only has exons, which contain the coding sequence.
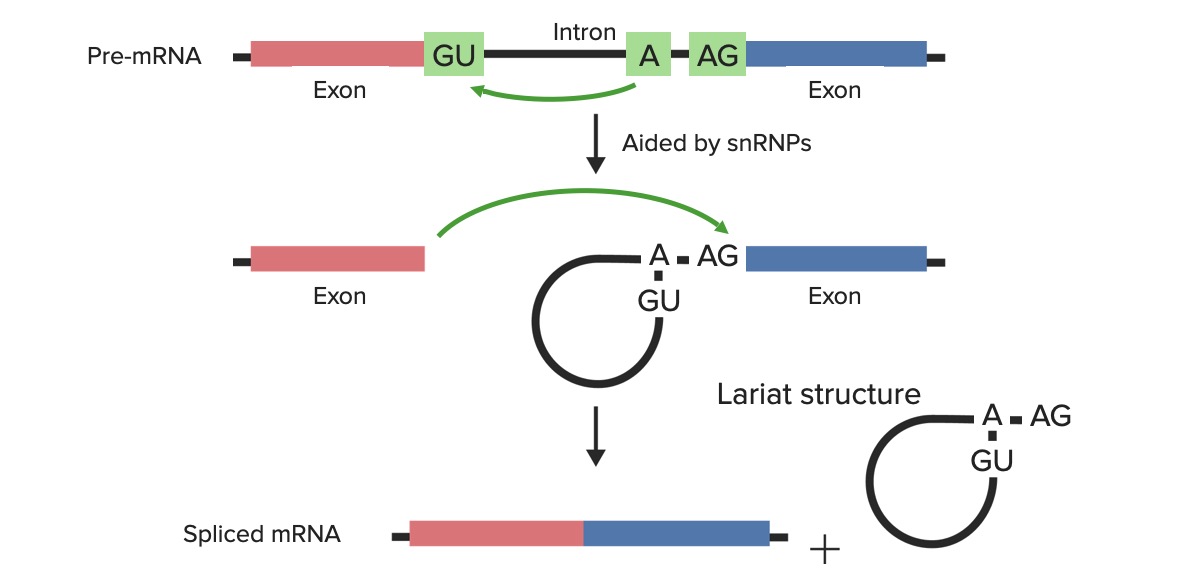
Technical aspects of splicing:
The pre-mRNA/hnRNA are made up of exons and introns. Small nuclear ribonucleoproteins + other proteins recognize the branch site and the exon–intron junctions where to cut: the 5’ donor site (containing the invariant GU sequence) and the 3’ acceptor site (containing the invariant AG sequence). The transcript of hnRNA + small nuclear ribonucleoproteins + other proteins combine at these sites and form the spliceosome.
Top image: Through the aid of small nuclear ribonucleoproteins (snRNPs), the first cut is made by the adenylyl residue (in the branch site) via a nucleophilic attack on the 5’ donor site.
Middle image: The free 5’ terminus then forms a bond with the branch site (making the lariat structure).
Bottom image: The second cut is made on the 3’ site of the intron and the exons are joined.
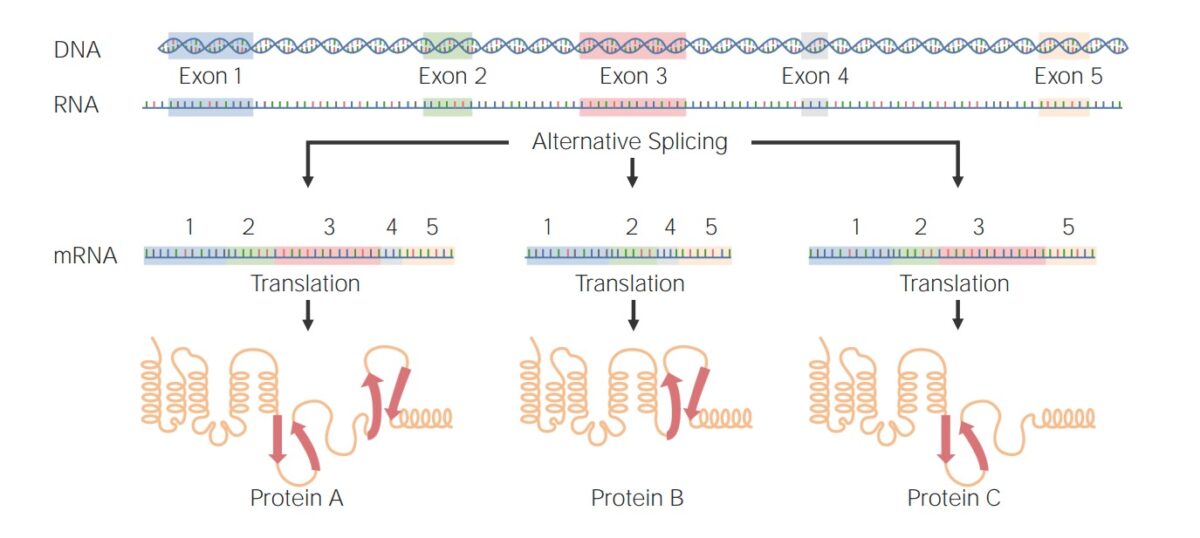
Examples of alternative splicing:
Protein A: Exons 1–5 were joined after splicing of introns.
Proteins B and C: An exon was selectively excluded to form a different protein.
Generally, the DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure sequence is reflected in the mature mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3′ end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure. Alteration of the sequence or RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure editing is an exception.
“C-to-U” editing:
“A-to-I” editing:
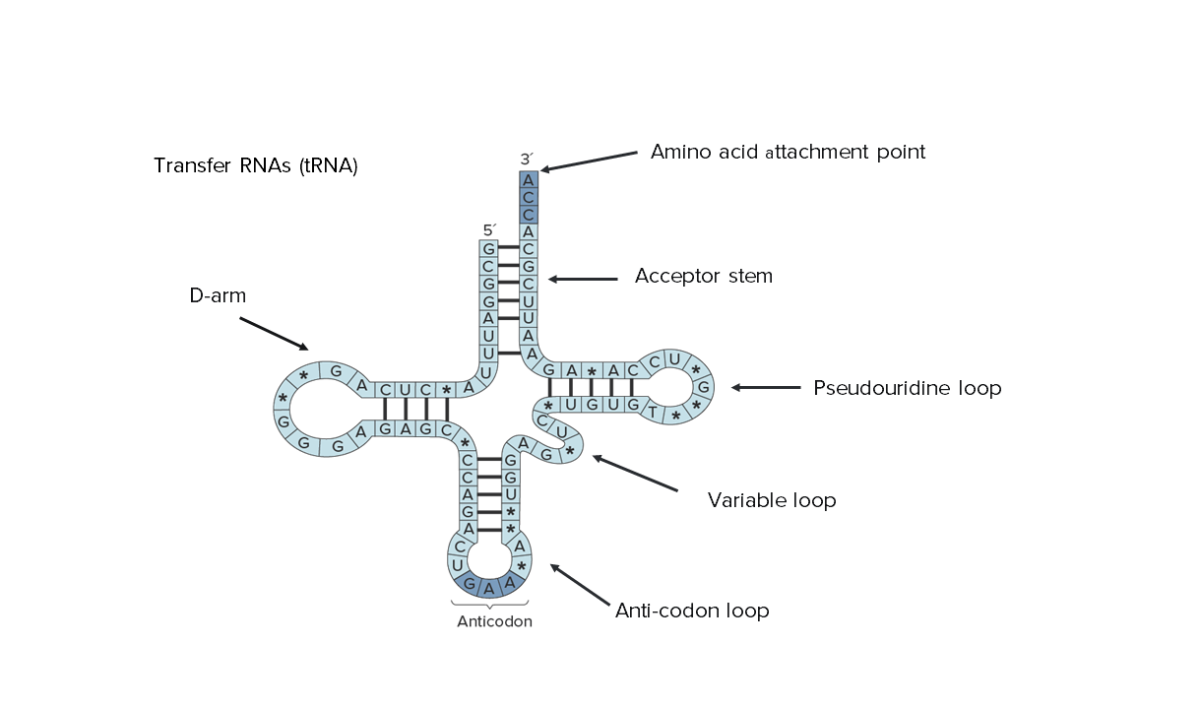
Secondary structure of transfer RNA (tRNA). Note that its entire sequence can be seen, indicating the reduced size.
Image by Lecturio.