Transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription is the process by which DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure is used as a template to make mRNA mRNA RNA sequences that serve as templates for protein synthesis. Bacterial mRNAs are generally primary transcripts in that they do not require post-transcriptional processing. Eukaryotic mRNA is synthesized in the nucleus and must be exported to the cytoplasm for translation. Most eukaryotic mRNAs have a sequence of polyadenylic acid at the 3' end, referred to as the poly(a) tail. The function of this tail is not known for certain, but it may play a role in the export of mature mRNA from the nucleus as well as in helping stabilize some mRNA molecules by retarding their degradation in the cytoplasm. RNA Types and Structure via an enzyme called RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure polymerase. Transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription is an important step in gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics expression, and as such, it is highly regulated. In prokaryotes, genes Genes A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. DNA Types and Structure are grouped together into DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure sequences, known as operons, that can be induced or repressed to regulate expression of these genes Genes A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. DNA Types and Structure together. Regulation in eukaryotes is much more complicated and involves a number of transcription factors Transcription Factors Endogenous substances, usually proteins, which are effective in the initiation, stimulation, or termination of the genetic transcription process. Stages of Transcription and regulatory sequences of DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure. Epigenetic mechanisms, including how the DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure is packaged, also play a role in transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription regulation by controlling what segments of DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure are available to the RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure polymerase.
Last updated: Jan 9, 2023
Definition:
DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure:
DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure is a double-helix molecule made up of 2 antiparallel strands. DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure has a structure that looks like a twisted ladder.
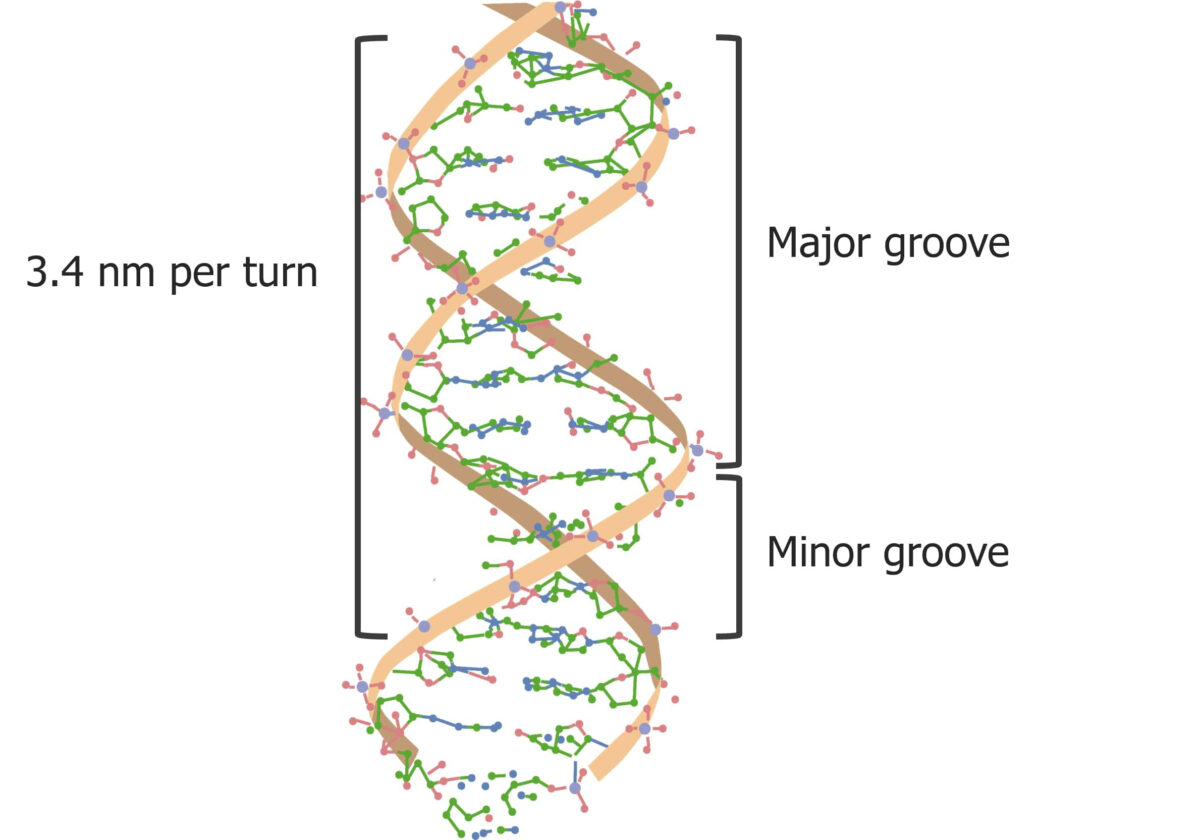
The major and minor grooves in DNA:
Most regulatory proteins bind to the major groove on the DNA helix. The major groove gives access to the nucleotide’s hydrogen bond donors.
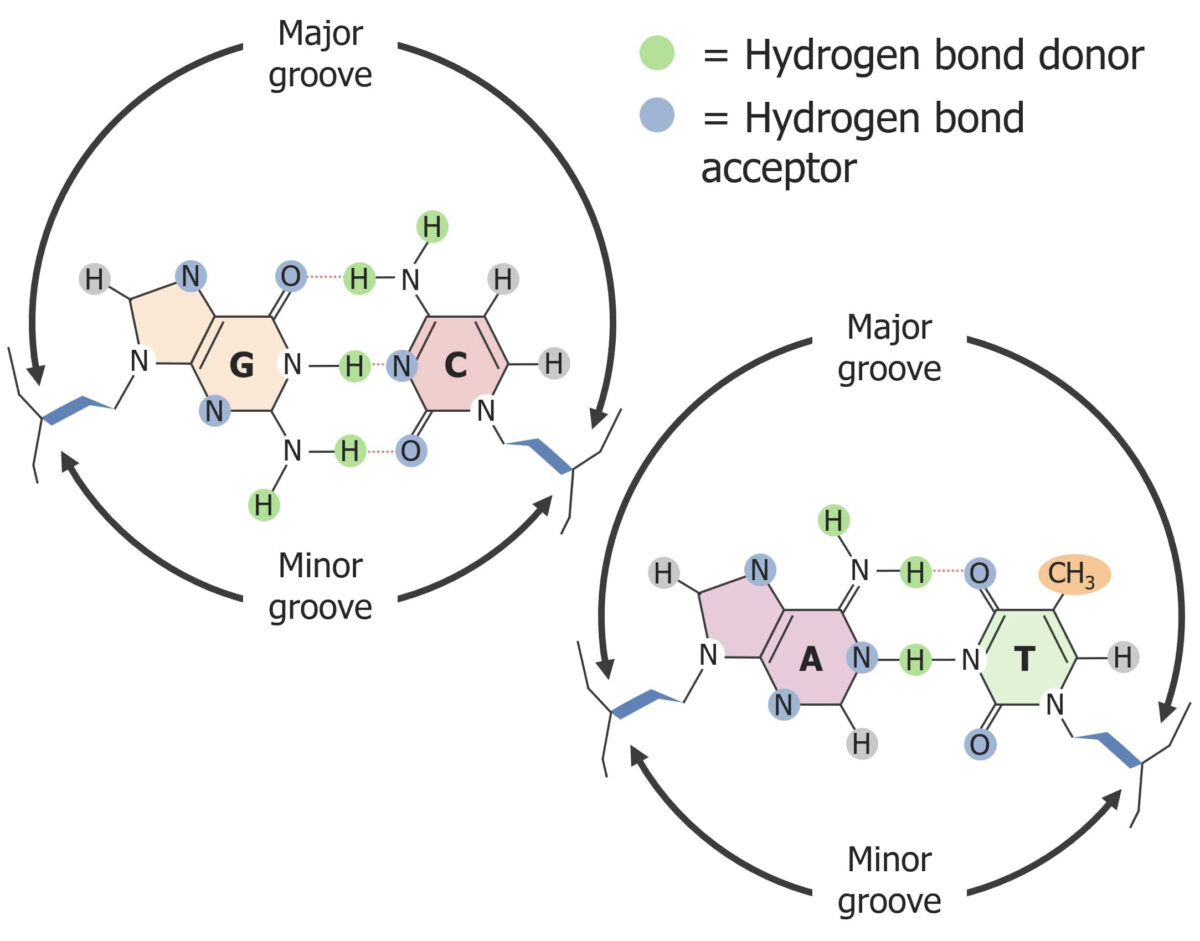
The major and minor grooves in DNA:
The major groove gives access to the nucleotide’s hydrogen bond donors.
RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure:
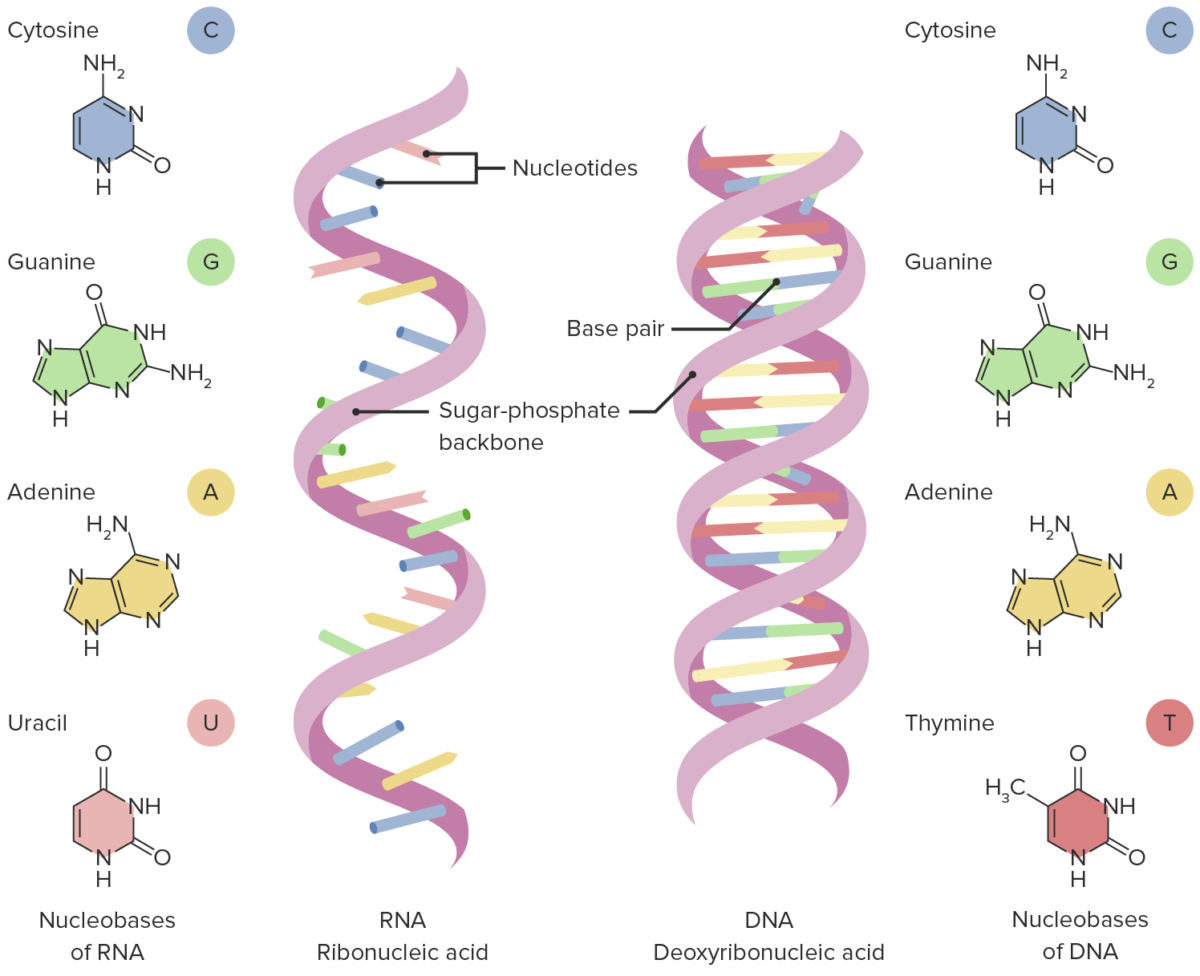
Structure of RNA and DNA
Image by Lecturio.Transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription regulation is mediated in part by regulatory proteins Regulatory proteins Proteins and Peptides that can bind BIND Hyperbilirubinemia of the Newborn to the DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure in its helical Helical Computed tomography where there is continuous x-ray exposure to the patient while being transported in a spiral or helical pattern through the beam of irradiation. This provides improved three-dimensional contrast and spatial resolution compared to conventional computed tomography, where data is obtained and computed from individual sequential exposures. Computed Tomography (CT) form (unwinding is not necessary) and regulate the transcriptional activity of RNA RNA A polynucleotide consisting essentially of chains with a repeating backbone of phosphate and ribose units to which nitrogenous bases are attached. RNA is unique among biological macromolecules in that it can encode genetic information, serve as an abundant structural component of cells, and also possesses catalytic activity. RNA Types and Structure polymerase.
Regulatory proteins Regulatory proteins Proteins and Peptides with the helix–turn–helix DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure binding motif have the following characteristics:
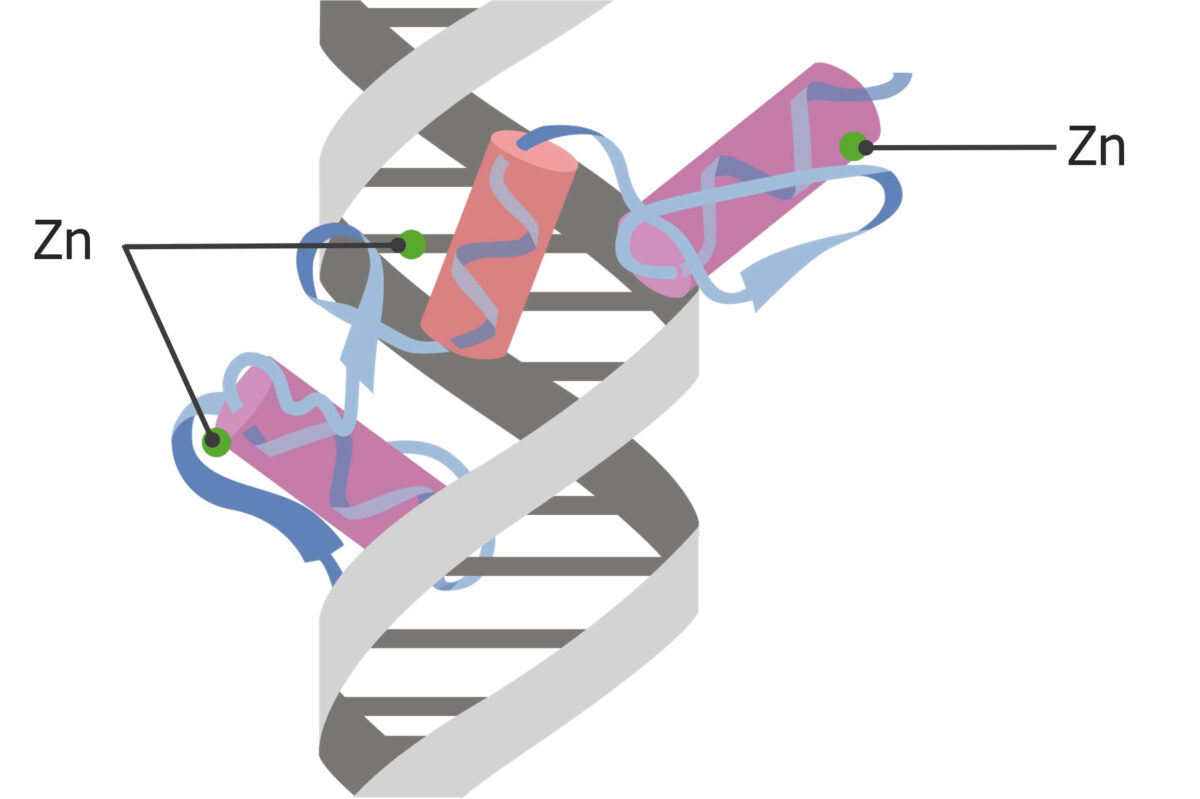
The zinc-finger binding motif:
Often used in steroid receptor binding mechanisms
Leucine zippers consist of 2 proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis, each containing a helical Helical Computed tomography where there is continuous x-ray exposure to the patient while being transported in a spiral or helical pattern through the beam of irradiation. This provides improved three-dimensional contrast and spatial resolution compared to conventional computed tomography, where data is obtained and computed from individual sequential exposures. Computed Tomography (CT) subunit and a hydrophobic subunit.
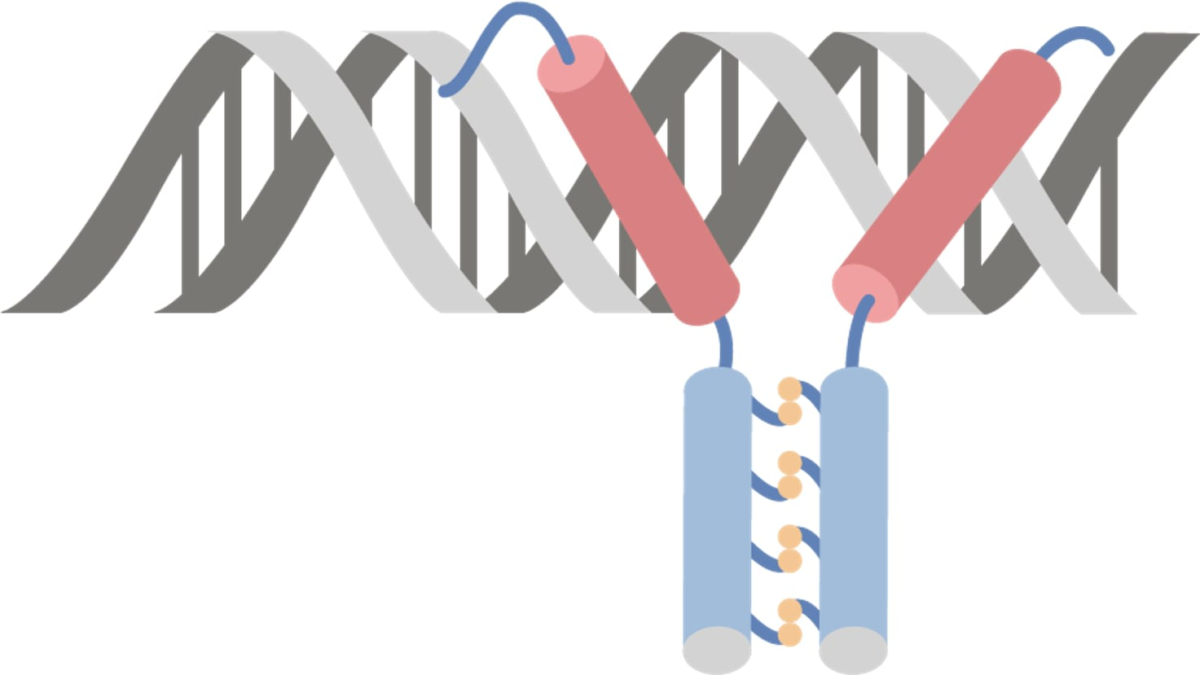
The leucine-zipper binding motif:
Has 2 subunits that zip together
Operons are clusters of coregulated genes Genes A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. DNA Types and Structure plus all the components that come together to regulate those genes Genes A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. DNA Types and Structure. Operons contain 2 primary regions of DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure sequences:
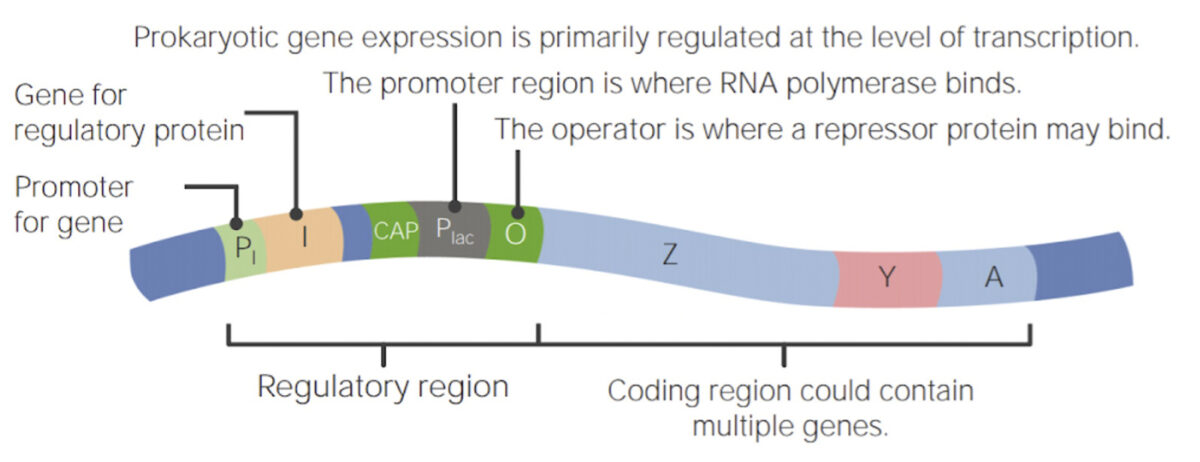
Example of a prokaryotic operon:
Prokaryotic gene expression is regulated primarily at the level of transcription.
Operons are clusters of coregulated genes in prokaryotes.
Inducible operons are operons that are off in the “normal” state and turn on under certain conditions. A common example is the lac operon in Escherichia coli Escherichia coli The gram-negative bacterium Escherichia coli is a key component of the human gut microbiota. Most strains of E. coli are avirulent, but occasionally they escape the GI tract, infecting the urinary tract and other sites. Less common strains of E. coli are able to cause disease within the GI tract, most commonly presenting as abdominal pain and diarrhea. Escherichia coli. Under normal circumstances, E. coli uses glucose Glucose A primary source of energy for living organisms. It is naturally occurring and is found in fruits and other parts of plants in its free state. It is used therapeutically in fluid and nutrient replacement. Lactose Intolerance for energy. When glucose Glucose A primary source of energy for living organisms. It is naturally occurring and is found in fruits and other parts of plants in its free state. It is used therapeutically in fluid and nutrient replacement. Lactose Intolerance is unavailable and/or lactose is present, the presence of lactose induces the lac operon:
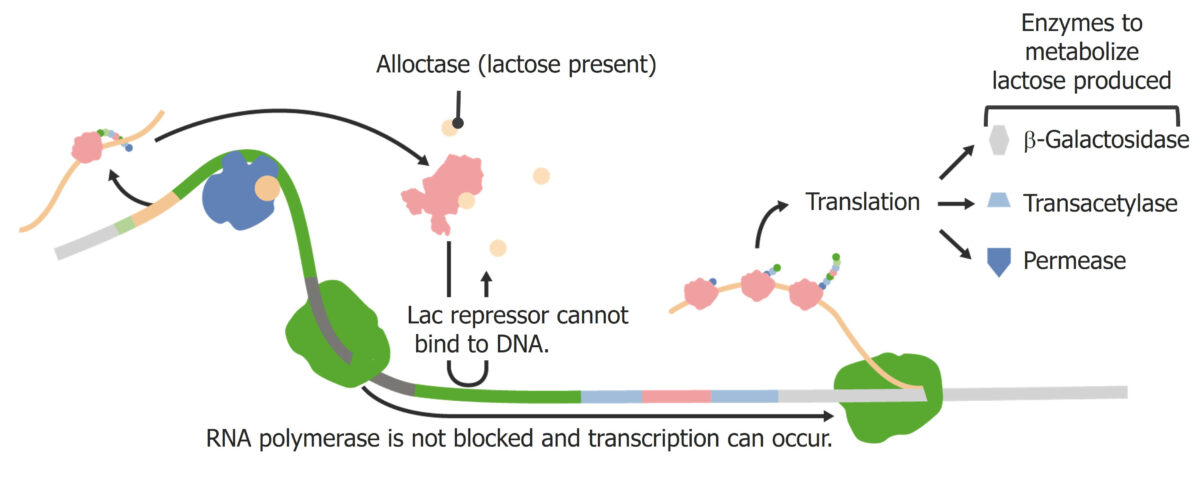
Lac operon:
In the presence of lactose, expression is activated.
Gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics regulation via the lac repressor:
Gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics regulation via the CAP activator:
Repressible operons are operons that are on in the “normal” state and turn off under certain conditions. The trp operon is a common example:
The default state is for the operon to be on and producing tryptophan.
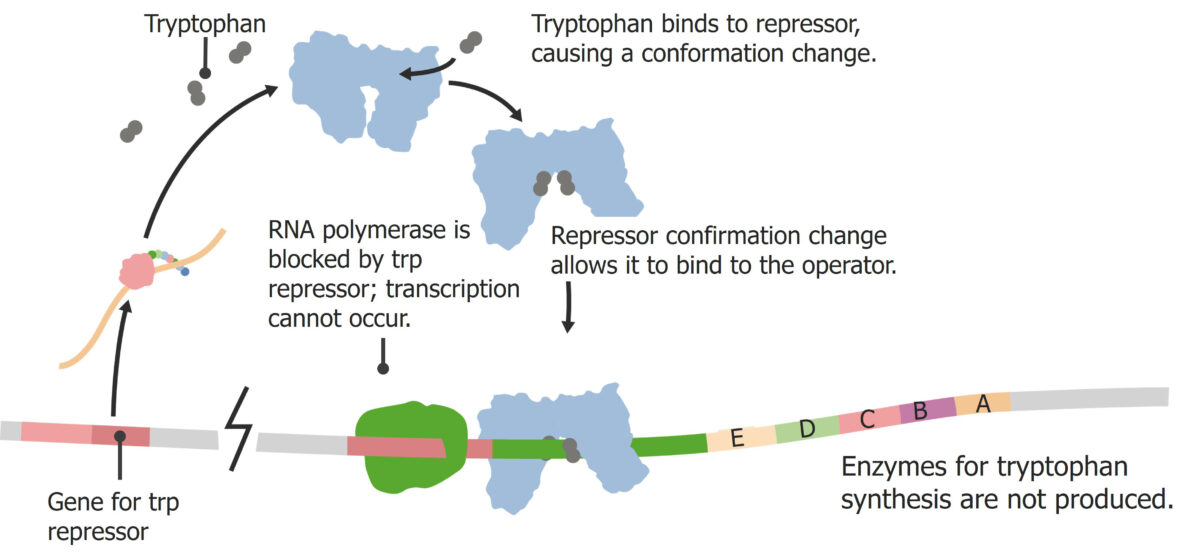
Trp operon:
Tryptophan activates the repressor.
Transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription factor (TF) is a generic term for proteins Proteins Linear polypeptides that are synthesized on ribosomes and may be further modified, crosslinked, cleaved, or assembled into complex proteins with several subunits. The specific sequence of amino acids determines the shape the polypeptide will take, during protein folding, and the function of the protein. Energy Homeostasis necessary for transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription. Each of these factors helps to regulate gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics expression.
Enhancer sequences of DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure help to initiate or augment transcription Transcription Transcription of genetic information is the first step in gene expression. Transcription is the process by which DNA is used as a template to make mRNA. This process is divided into 3 stages: initiation, elongation, and termination. Stages of Transcription, which further promotes gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics expression.
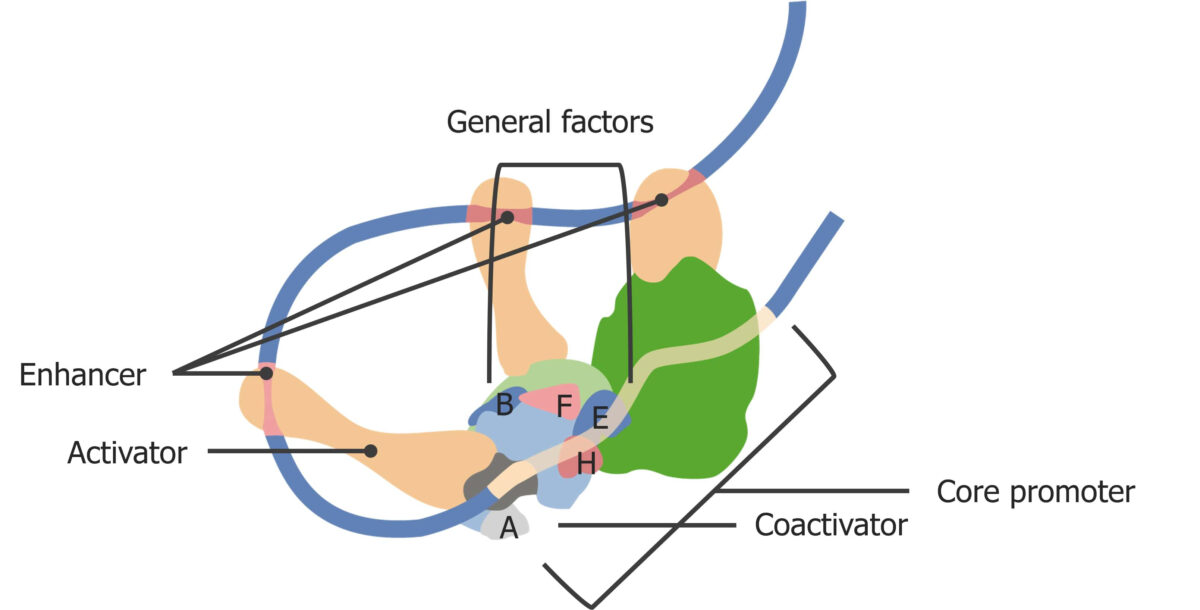
Eukaryotic transcription initiation complex
Image by Lecturio.Epigenetic regulation Epigenetic regulation Epigenetic regulation is regulation of gene expression that does not involve alterations to the DNA sequence or any of its transcribed products. The most common forms of epigenetic regulation are DNA methylation, which suppresses gene expression, and modifications to the histone proteins, which affect the structure of DNA packaging. Epigenetic Regulation is the regulation of gene Gene A category of nucleic acid sequences that function as units of heredity and which code for the basic instructions for the development, reproduction, and maintenance of organisms. Basic Terms of Genetics expression that does not involve alterations to the DNA DNA A deoxyribonucleotide polymer that is the primary genetic material of all cells. Eukaryotic and prokaryotic organisms normally contain DNA in a double-stranded state, yet several important biological processes transiently involve single-stranded regions. DNA, which consists of a polysugar-phosphate backbone possessing projections of purines (adenine and guanine) and pyrimidines (thymine and cytosine), forms a double helix that is held together by hydrogen bonds between these purines and pyrimidines (adenine to thymine and guanine to cytosine). DNA Types and Structure sequence or any of its transcribed products. Epigenetics include:
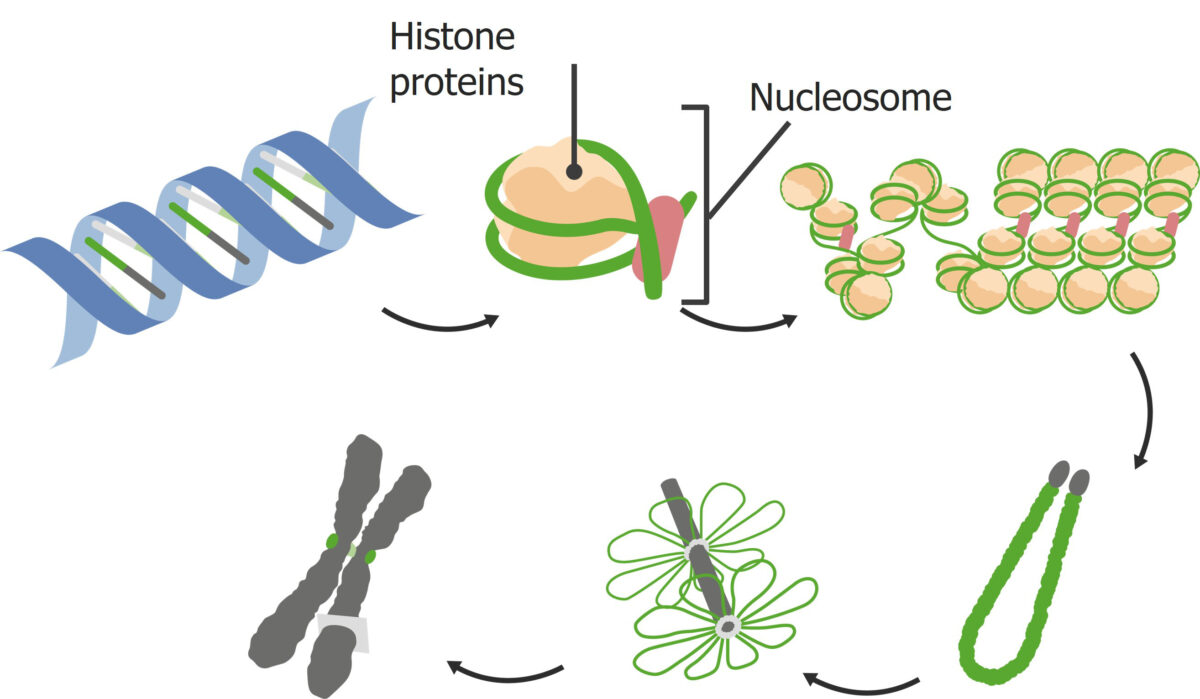
DNA packing:
How DNA is packaged into nucleosomes and then chromosomes.
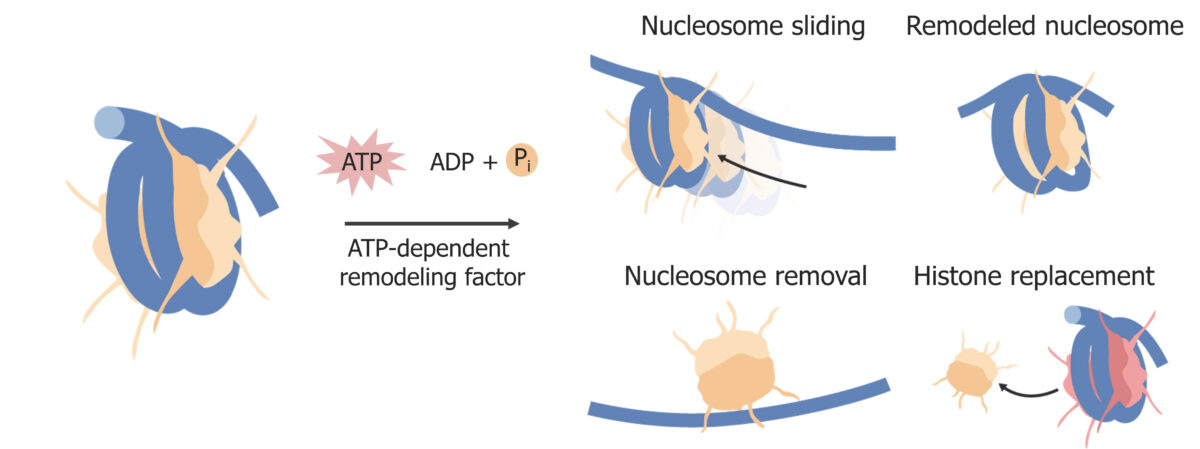
Nucleosome remodeling:
Examples of changes to histones. Chromatin remodeling factors also alter chromatin structure. Chromatin uses energy from ATP.