Playlist
Show Playlist
Hide Playlist
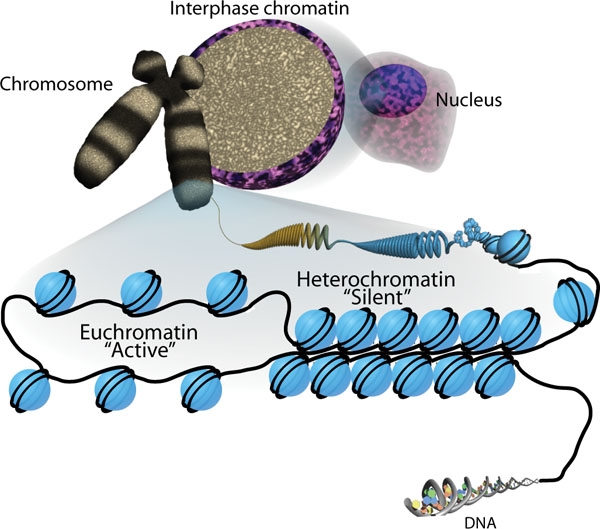
Prokaryotic Replication – DNA Structure and Replication
-
Slides 05 DNAStructureAndReplication Genetics.pdf
-
Reference List Molecular and Cell Biology.pdf
-
Download Lecture Overview
00:00 Nun müssen wir verstehen, wie die Replikation abläuft. Was ist das allgemeine Muster und dann werden wir ins Detail gehen. Zunächst werden wir wir mit einem Blick auf prokaryotische Replikation beginnen. Bei der prokaryotischen Replikation ist es so, Sie erinnern sich, dass die Bakterien ein einziges zirkuläres Chromosom besitzen. Es sieht vielleicht nicht wie ein Kreis aus, es könnte im Zellkern ganz zerbröselt sein, aber es ist einzeln und kreisförmig und daher hat jedes von ihnen einen spezifischen Ursprung bei der Replikation und eine bestimmte Sequenz, die die Beendigung der Replikation besagt. 00:36 In diesem bakteriellen Chromosom werden wir ein geöffnetes Replikon oder einen geöffneten Raum sehen, in denen wir unsere Enzyme oder Enzymmaschinen haben. Wir nennen diese Maschinerie ein Replisom und diese Replisomen bewegen sich in beide Richtungen um die zirkulären Chromosomen herum bis sie beide an den Terminus gelangen, in diesem Fall werden sich die zwei neuen Chromosome voneinander trennen und in ihre jeweiligen Zellen gehen. Ein ziemlich einfacher Vorgang, oder? Nun, nicht ganz. 01:12 Es ist die Maschinerie, die sich in den beiden Replikonen im Replisom abspielt, die also an dem ganzen Replikationsprozess beteiligt ist, die wir in dieser Vorlesung kennenlernen werden. Auch hier muss ich es wiederholen, weil es immer wieder auftaucht, die DNA-Polymerase ist das wichtigste Enzym, die sich an Nukleotiden festsetzt, kann nur in Richtung 3' zu 5' lesen. 01:40 Die andere wichtige Information ist, dass die DNA-Polymerase auch diese 3' OH-Gruppe braucht. 01:47 Diese 3'-OH-Gruppe, die ich immer wieder betone, ist wirklich wichtig zu kennen. 01:51 Es braucht eine dreipolige OH-Gruppe, um sich an das vorherige Nukleotid zu hängen und sich an das nächste Nukleotid anzuhängen. Nehmen wir an, wir betrachten den Schablonenstrang und sehen uns den neu synthetisierten Strang an. Wir werden sagen, dass das 3'-Ende sich auf diesem Templatestrang befindet und es in dieser Richtung weiter geht. Die DNA-Polymerase muss den Template-Strang in diese Richtung lesen, aber es legt einen komplementären Strang in dieser Richtung an. 02:22 Wir haben also ein Nukleotid mit dem 3'-OH an diesem Ende und ein weiteres Nukleotid mit 3'-OH, das an diesem Ende heraushängt. Der neu synthetisierte Strang wird nun in der Richtung 5'n zu 3' orientiert sein. Das 3'-OH ist entscheidend beim Ablesen, immer 3' zu 5', das Anlegen neuer Nukleotide erfolgt 5' zu 3' erstklassig. Das ist wichtig, um zu verstehen, warum die DNA-Replikation ziemlich verwirrend sein kann. 03:00 Schauen wir uns mehr Details an. Hier ist unser Nukleotid. Wir wollen das Stück mit dem Zucker betonen. Wir haben unsere 3'-OH und unsere 5'-Phosphate. Super wichtige Teile des Moleküls, um zu verstehen, wie sie alle im neusynthetisierten DNA-Strang zusammengesetzt sind. 03:22 Diese Elternstränge werden antiparallel sein. Wir haben eine Replikationsgabel. 03:28 Wenn sich diese Replikationsgabel öffnet, können wir sehen, dass die Template-Stränge in gewisser Weise in entgegengesetzter Richtung liegen. Wir haben einen führenden Strang, der von 3', das grüne Stück, von 3' zu 5' ausgerichtet ist, überhaupt kein Problem. Die DNA-Polymerase kann entlang dieses Strangs laufen und kontinuierlich Nukleotide anlegen. Der andere Strang jedoch, der alte komplimentäre Strang, hat ein kleines Problem, denn jetzt können wir bei der Replikationsgabel nur lesen, was oben als 5'-Ende herausragt, und der grüne Strang geht zum 3'-Ende. In diesem Fall hat die DNA- Polymerase ein Problem, da sie nicht von 5' zu 3' lesen kann. Sie muss den grünen Strang von 3' zu 5' lesen und die Nukleotide antiparallel ablegen. Das erfordert also ein wenig Zurückspringen. 04:33 Wir werden uns gleich die Details ansehen, wie das funktioniert, aber wenn wir zurückspringen, benötigen wir verschiedene Fragmente. Wir nennen diese Fragmente Okazaki-Fragmente. Ich werde Ihnen kurz zeigen, wie wir mit diesen umgehen. Okazaki-Fragment ist ein Wort, das Sie kennen müssen. Merken Sie sich, dass die Replikation bidirektional ist. Es gibt eine Replikationsgabel an beiden Enden. Über eine Seite haben wir gesprochen. Hier haben wir einen nacheilenden Strang oben, einen führenden Strang unten. 05:03 Am anderen Ende wird der nacheilende Strang auf der Unterseite und der führende Strang oben sein, aber auf diesem unteren Strang, den wir jetzt lesen,ist die Vorlage von 5' zu 3', was für die DNA-Polymerase nicht funktioniert. Wir benötigen Okazaki-Fragmente auf diesen unteren Strängen, es findet also eine bidirektionale Replikation statt.
About the Lecture
The lecture Prokaryotic Replication – DNA Structure and Replication by Georgina Cornwall, PhD is from the course Understanding Genetics.
Included Quiz Questions
DNA replication always proceeds by the addition of new bases to which end of an existing strand?
- 3'
- 1'
- 2'
- 4'
- 5'
Where are Okazaki fragments used during replication?
- The lagging strand of the chromosome
- The leading strand of the chromosome
- The sense strand of the chromosome
- The nonsense strand of the chromosome
- The missense strand of the chromosome
Which of the following terms best describes DNA replication in prokaryotes?
- Bidirectional
- Unidirectional
- Tridirectional
- Unilateral
- Bilateral
Customer reviews
4,0 of 5 stars
| 5 Stars |
|
0 |
| 4 Stars |
|
1 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
4/5 also add on the replication bubble but as far as the other information well done i understood the concept