Playlist
Show Playlist
Hide Playlist
Post-transcriptional Modification: Splicing
-
Slides 07 TranscriptionProkaryotesEukaryotes Genetics.pdf
-
Reference List Molecular and Cell Biology.pdf
-
Download Lecture Overview
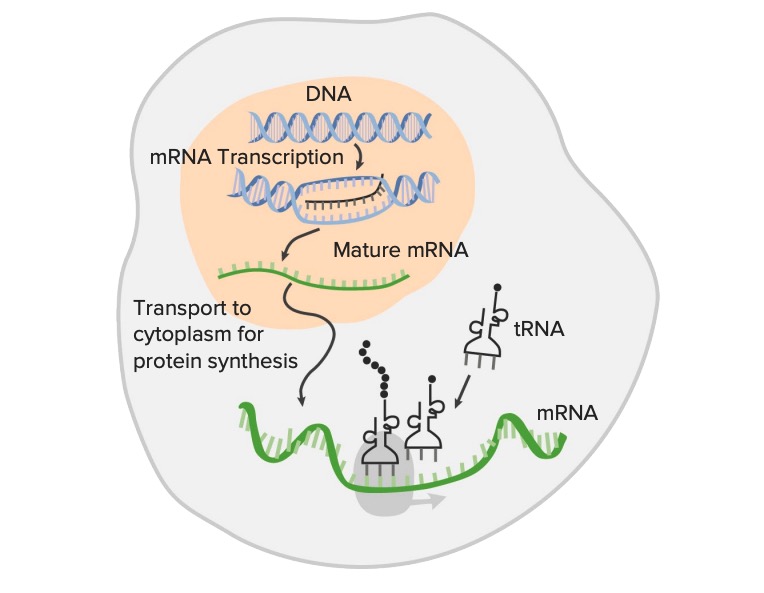
00:00 Now, at the same time as this is happening, we have another series of things happening, which is called RNA splicing. 00:06 The primary RNA transcript from eukaryotic cells contains information that is not coding. 00:16 So we have it split up into introns and exons. 00:20 And they’re not as neat and symmetrical as these, but these will exhibit the whole concept. 00:27 Introns are inactive and exons are expressed. 00:33 At least, that’s the way I remember them because it’s not truly true that introns are inactive. 00:38 But exons are expressed. 00:40 They’re the DNA that’s going to end up in the mature mRNA. 00:43 So we cut out the introns and produce a mature mRNA and you have a 5’ cap and a poly-A tail. 00:52 So the way that the cell manages cutting out the introns is through the use of a tool called a spliceosomes. 00:59 The spliceosome is like a pair of giant molecular scissors and it is composed of snRNPs, S-N-R-N-Ps, which are small nuclear ribonuclear proteins. 01:12 Try saying that one. 01:14 That’s why we call them snRNPs because it’s much easier. 01:17 So these small nuclear RNA portion of the snRNP is going to base pair with the 5’ end of the intron as well as the branch site of the intron. 01:31 You’ll see how that comes into play shortly. 01:32 And we’ll also see some bind with the 3’ end. 01:36 They come together to form a whole spliceosome and then they cut the DNA and stick one piece, the Lariat loop, back onto the branch site and then cut the 3’ end and excise the intron. 01:53 So the intron is removed. 01:56 Now the two exons can be stuck together and we are good to go with introns being removed. 02:03 But let’s go a little bit further and look at the possibilities because we’ve now learned that we can have alternative splicing. 02:14 So, yes, we have introns and exons and exons are the things that are going to be expressed. 02:20 But the pieces that you choose to be expressed or the cell chooses to be expressed could be different depending on how the spliceosomes cut. 02:33 So we could out introns in this fashion and produce a protein that looks like protein A or we could cut it in this fashion and stick those pieces together as exons and have them express a different protein. 02:47 And then we could have a different variety even still, so alternative splicing ends up with the production of a variety of different proteins from the same messenger RNA. 03:02 Now, it sort of seems like this might be violating that one gene, one polypeptide thing. 03:08 I mean, you could argue that it’s still one polypeptide. 03:12 But as definitely seeing that the protein comes out in a little different way, but this is one of the ways that we have with so few genes, we have so much possibility for a variation. 03:25 It’s these mechanisms of alternative splicing. 03:27 So we can one coding sequence, actually results in multiple possible proteins. 03:35 So you may have heard that we got done early sequencing the human genome back in the year 2000. 03:42 And part of the reason was it turned out that we had a lot less genes than we had anticipated. 03:48 For example, we have much less than corn, which was quite confusing to us. 03:53 Because apparently, we are more complex than corn and this alternative splicing is perhaps one of the explanations for why we could be more complex because we have the possibility of multiple protein products from one coding sequence of DNA. 04:09 So let’s recap what’s happened in this RNA processing phase. 04:14 We had to take our primary RNA transcript and make it into mature messenger RNA. 04:20 We added a 5’ cap to protect him from the environment out there in the cytosol as well as help him bind the ribosomes. 04:29 In addition to that, we added a poly-A tail to protect him from enzymes that might be out there trying to degrade the messenger RNA. 04:38 And then we cut out introns in order to make a mature messenger RNA that’s ready to go out into the cytosol and be translated into a polypeptide by ribosomes out there. 04:52 That is precisely the topic of our next lecture. 04:55 So I look forward to seeing you for that one. 04:57 At this point though, you should be able to describe the process of transcription in prokaryotes as well as differentiate between the initiation phase, the elongation phase, and termination phase, and explain differences in prokaryotic and eukaryotic transcription as well diagram some of the aspects of RNA processing. 05:21 Thank you so much for your attention. I will see you shortly.
About the Lecture
The lecture Post-transcriptional Modification: Splicing by Georgina Cornwall, PhD is from the course Gene Expression.
Included Quiz Questions
You are characterizing a novel protein in mice. Analysis shows that high levels of the primary transcript coding for this protein are found in the brain, muscle, liver, and pancreatic tissues. However, an antibody that recognizes the C-terminal portion of the protein indicates that the protein is present in the brain, muscle, and liver, but not in the pancreas. What is the most likely explanation for this result?
- Alternative splicing in the pancreas yields a protein that is missing the portion that the antibody recognizes.
- The gene that codes for this protein is not transcribed in the pancreas.
- There is no modification of the primary transcript in the pancreas.
- Alternative splicing in the brain, muscles, and liver decreases the level of translation.
- Introns of the primary transcript were spliced together instead of the exons in the pancreatic tissues.
Which of the following is CORRECT regarding spliceosomes? Select all that apply.
- Spliceosomes act like molecular scissors to cut the introns from the pre-mRNA transcript to generate mature mRNA.
- Spliceosomes are composed of snRNPs.
- Spliceosomes add a 5' cap and poly-A tail to pre-mRNA.
- Spliceosomes help in the transfer of mRNA from the transcription bubble to the exterior of the nucleus.
- Spliceosomes excise exons from tRNA and join introns together to generate a fully functional tRNA molecule.
Which of the following is best explained by alternative splicing?
- The genetic diversity in eukaryotes
- The genetic similarity between eukaryotes and prokaryotes
- The genetic similarity among eukaryotes
- The genetic similarity among prokaryotes, eukaryotes, and viruses
- The genetic similarity among prokaryotes
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
5 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |