Playlist
Show Playlist
Hide Playlist
Negative-sense Single-stranded RNA ((-)ssRNA) Virus – RNA Virus Genomes
-
Slides 05 ViralGenomes MicrobiologyAdvanced.pdf
-
Download Lecture Overview
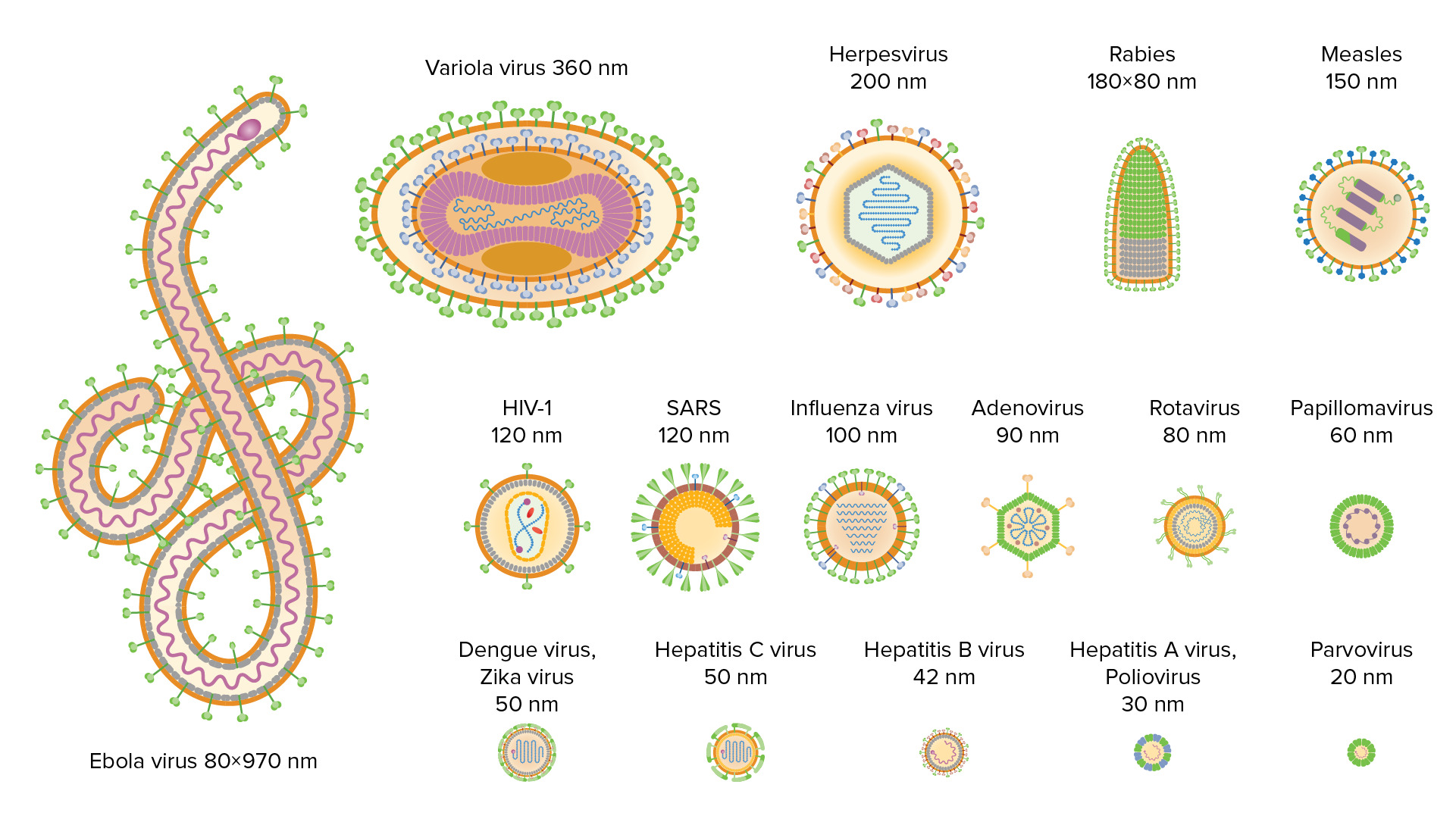
00:00 Let's go to the minus polarity single-stranded RNAs. There are a number of important human viruses here. The Paramyxoviruses including measles virus, and mumps virus, important childhood infections, completely controllable by vaccination. Rhabdoviridae, these contain rabies virus, the infection transmitted by rabid dogs. The Filoviruses, everyone should know about the filoviruses now, because it was just a big outbreak of Ebola virus infection in Western Africa. The filoviruses are unusual filamentous particles; they include not only the Ebola viruses but Marburg viruses first discovered in Germany at a primate facility. 00:46 The Orthomyxoviridae includes the very important human pathogen, influenza virus, which causes annual outbreaks of respiratory disease in many, many millions of people. And finally the Arenaviridae, which Lassa virus is the best-known. This is a virus that also causes a hemorrhagic fever, largely in Africa. These viruses have a negative sense RNA genome, and from what I've told you so far, you should be able to predict exactly what is going to happen in this flow of genetic information. Minus strand RNA cannot be translated, and the cell cannot make a copy of it to make messenger RNA so you can predict the answer to the question: Do negative stranded RNA viruses carry an RNA polymerase in the particle? The answer of course is yes, because it has to come in with the RNA and then copy it to make plus stranded RNA. If there were no polymerase in the particle, that would be the end of the infection, because the negative RNA can't be translated, it can't be copied by any cell enzyme; it would be the end of the infection. So these genomes have evolved to include the RNA polymerase in the particle. So plus stranded RNA can be made, messenger RNA can be translated into proteins, and also plus stranded RNAs can be made to make more genomes, to make negative RNA genomes, which get incorporated into the particles. 02:15 There are two kinds of viruses in general with negative stranded RNA genomes. We have some like the influenza viruses, where the genome is segmented; it's in pieces like the double-stranded RNA genomes of the real viruses. So you can see here there are eight negative strand RNA segments in the influenza viruses. Then there is another kind of negative strand RNA virus where the genome is not segmented. It exists in the virus particle as a long RNA for the Paramyxoviruses, measles and mumps 15 to 16 kb, for the rabies like viruses 13 to 16 kb. Again these are all negative strand viruses, two general ways of assembling them in the virus particle. 03:02 Having a segmented genome, the influenza virus, the reoviruses, and many others is very important for genetic variability, because of this example. When you infect a cell with one of these viruses, the RNA is replicated in the cell and you have many, many copies of the RNA which are then packaged into new virions. If you would by chance infect a cell with two different viruses, here we’re showing two different influenza virus labeled L and M, infecting the same cell, if they both infect the cell, their RNAs are all going to be replicated at the same time. They're all mixed together, and the new viruses that are produced are not only going to be L and M viruses, but recombinants that have RNA segments from either parent, and one of the recombinants is shown here. It's called R3 and you can see that second RNA segment is red, it derives from the red parent and the rest of them are from the blue parent. In fact you could get almost any reassortant possible out of this co-infection and this is partly the reason why influenza viruses are problematic. They vary extensively and this is one of the reasons. This kind of variation by reassortment, is why we generate new influenza viruses on a daily basis and why every 30 to 40 years, we have a brand-new strain that causes a global pandemic, because it's a reassortant caused by this process.
About the Lecture
The lecture Negative-sense Single-stranded RNA ((-)ssRNA) Virus – RNA Virus Genomes by Vincent Racaniello, PhD is from the course Viruses.
Included Quiz Questions
Which of the following statements regarding viral RNA genomes is correct?
- Positive-sense single-stranded RNA genomes may be translated directly by the host cell ribosomes to make viral protein.
- Double-stranded RNA genomes can be directly translated to make viral protein by ribosomes.
- The positive-sense RNA strand of a double-stranded RNA virus has the sole purpose of serving as a template for viral replication.
- RNA genomes can be copied by host cell RNA-dependent RNA polymerases.
- Positive-sense single-stranded RNA virus replication cycles do not require a negative-strand intermediate.
Ebolaviruses belong to which of the following families?
- Filoviridae
- Paramyxoviruses
- Orthomyxoviridae
- Arenaviridae
- Mumps viruses
The presence of which enzyme is essential for the synthesis of viral proteins by Orthomyxoviridae?
- Polymerase
- Mitochondria
- Lysosome
- Golgi apparatus
- Endoplasmic reticulum
New strains of influenza arise frequently because of which of the following processes?
- Reassortment
- Loss of cellular energy
- Extensive replication
- Misfolding of viral proteins
- Genetic breakdown
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
1 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
the explanation is very clear, the scientific names are pronounced very well and the illustrations are standing out.