Playlist
Show Playlist
Hide Playlist
Elongation of mRNA
-
Slides 07 TranscriptionProkaryotesEukaryotes Genetics.pdf
-
Reference List Molecular and Cell Biology.pdf
-
Download Lecture Overview
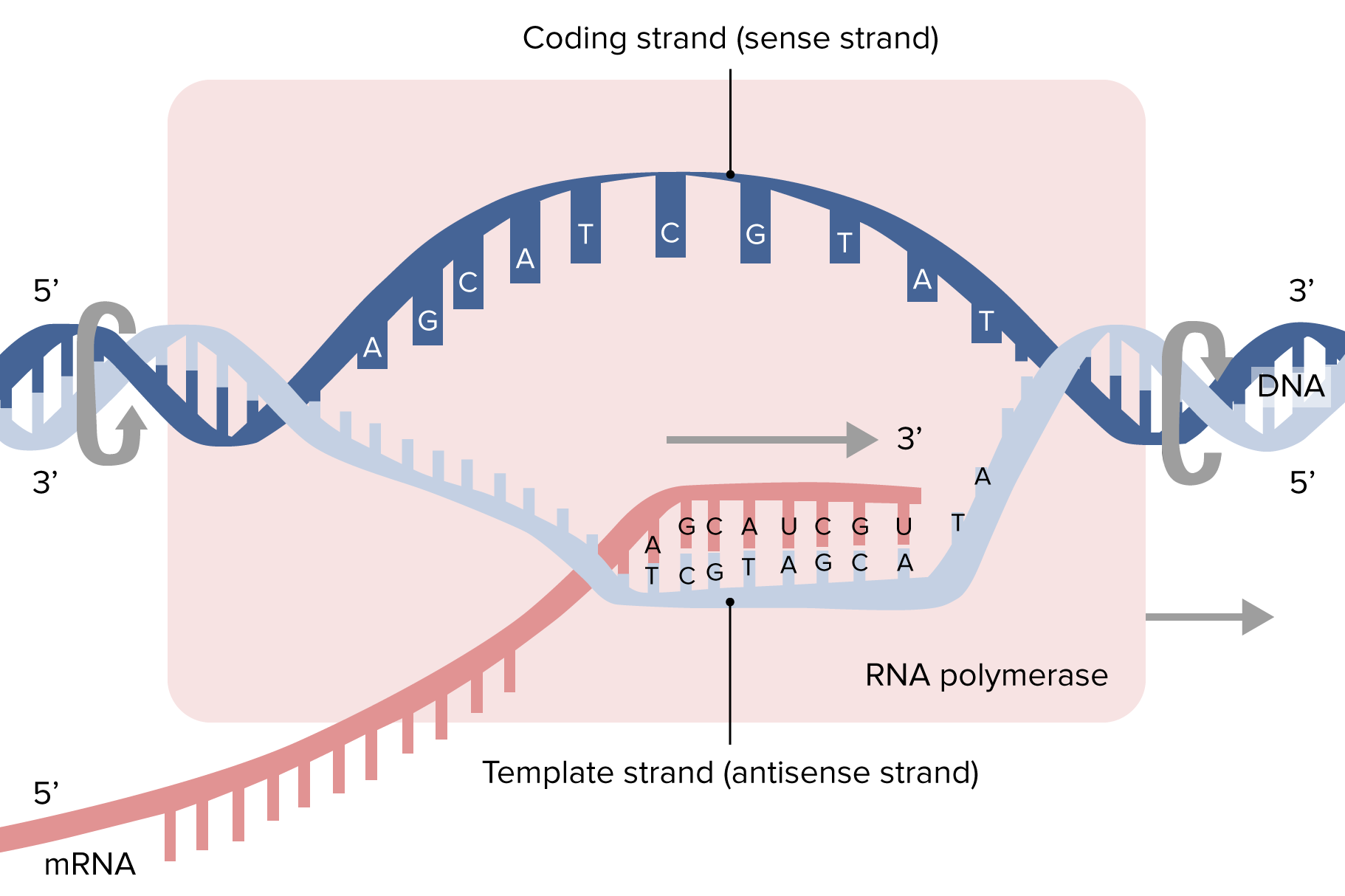
00:01 Nun muss der wachsende Strang verlängert werden. 00:04 Kehren wir zurück zur einfachen Darstellung der Polymerase. 00:08 Die RNA-Polymerase bildet eine Transkriptionsblase. 00:16 In der Transkriptionsblase sind die getrennten DNA-Einzelstränge und die synthetisierte messenger-DNA. 00:23 Genau wie die DNA-Polymerase liest die RNA-Polymerase den Matrizenstrang von 3' nach 5' ab und bildet ein messenger-RNA Transkript, das von 5' nach 3' verläuft. 00:38 Der Beginn des Transkripts ist dessen 5'-Ende. 00:43 Die Polymerase fügt Nukleotide an die wachsende Nukleotidkette. Dabei wird U statt T verwendet. 00:51 Genau wie bei der DNA-Replikation handelt es sich um Nukleosidtriphosphate. 00:59 Alle Nukleotide, die polymerisiert werden, bringen also ihrer eigene Energie für die Reaktion mit. 01:04 Die Nukleosidtriphosphate liegen als einzelne Nukleotide vor. 01:08 Zwei Phosphate werden abgespalten, wodurch die benötigte Energie für die Katalyse zur Verfügung gestellt wird. 01:13 Das ist genau wie bei der Replikation. 01:16 Ich weiß nicht mehr, ob ich bei der Replikation den Triphosphat-Teil der Nukleotide erwähnt habe, beachten Sie also, dass dieses Prinzip für beide Prozesse gilt. 01:25 Sobald die Elongationszyklen beendet sind, die Sequenz durch Nukleotide vervollständigt und unsere messenger-RNA hergestellt wurde, muss sich die messenger-RNA von der DNA lösen. 01:40 Das passiert, wenn die Terminator-Sequenz erreicht wird. 01:43 Diese Terminator-Sequenzen sind interessant. 01:47 Es handelt sich um ein Sequenz bestehend aus G- und C-Basenpaaren. 01:49 Die Basenpaare G und C haben eine ziemlich starke Assoziation zueinander. 01:55 Das führt zur Bildung einer Haarnadelschleife im Transkript der messenger-RNA. 02:01 Nach diesen G- und C-Paarungen gibt es einige A- und T-Paarungen, und dann folgt eine Reihe von Uracil. 02:15 Die endständige Uracil Sequenz ist ziemlich schwach, da Wasserstoffbrückenbindungen zwischen den U-A-Basenpaaren leicht aufgelöst werden können. 02:28 Dadurch fällt die messenger-RNA aus dem Holoenzym heraus und wird freigesetzt. 02:37 Jetzt haben wir ein fertiges Transkript der DNA in Form einer messenger-RNA.
About the Lecture
The lecture Elongation of mRNA by Georgina Cornwall, PhD is from the course Gene Expression.
Included Quiz Questions
Which of the following describes the transcription terminator sequence on the coding strand of DNA?
- The presence of a 20-base-pair GC-rich region followed by a small poly-T stretch
- The presence of a small poly-T stretch followed by a 20-base-pair GC-rich region
- The presence of a small poly-U stretch followed by a 20-base-pair AT-rich region
- The presence of a 20-base-pair AT-rich region followed by a small poly-T stretch
- The presence of a 20-base-pair AC-rich region followed by a small poly-UG stretch
Which of the following best describes the elongation of mRNA in prokaryotes?
- The addition of ribonucleotides by RNA polymerase in a 5' to 3' orientation
- The addition of deoxyribonucleotides by RNA polymerase in a 5' to 3' orientation
- The addition of ribonucleotides by RNA polymerase in a 3' to 5' orientation
- The addition of ribonucleotides by DNA polymerase in a 5' to 3' orientation
- The addition of deoxyribonucleotides by RNA polymerase in a 3' to 5' orientation
Customer reviews
1,5 of 5 stars
| 5 Stars |
|
0 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
1 |
| 1 Star |
|
1 |
More organization is needed to explain this lecture because is an important part of the DNA synthesis.
The instructor reading off the podium is not appreciated. It doesn't entice the viewer to keep watching.