Playlist
Show Playlist
Hide Playlist
DNA Amplification – Analytical Techniques in Biotechnology
-
13 Advanced BiotechnologyAnalyticalTechniques.pdf
-
Reference List Biochemistry.pdf
-
Download Lecture Overview
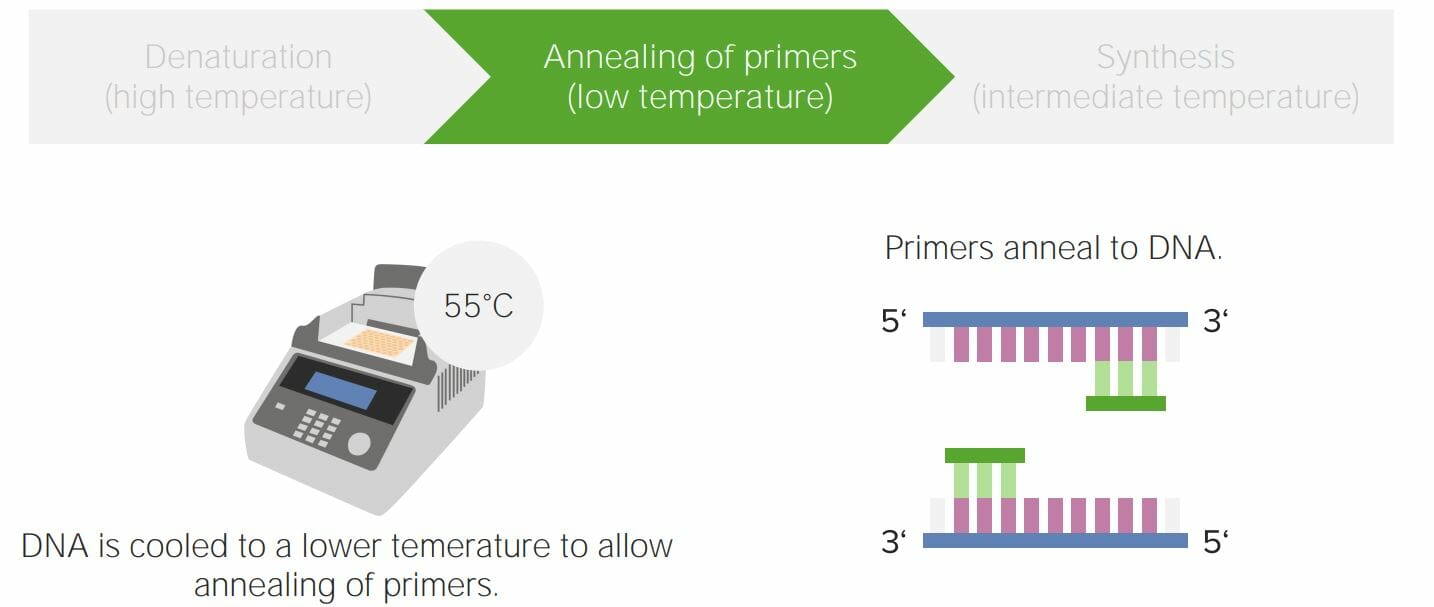
00:01 Man sagt, dass Wissen Macht ist, und mit Biotechnologie können wir in dieser Präsentation sehen, warum es wahr ist, denn das Wissen über molekulare biologische Prozesse dient zur Entwicklung leistungsfähiger Analysetechniken. 00:13 Ich werde in diesem Vortrag über drei dieser Techniken sprechen. 00:16 Diese sind DNA-Amplifikation, Microarray-Analyse Analyse und 2D-Gel-Analyse. 00:21 Nun das Wissen über den Prozess der DNA-Amplifikation wurde in einer Technik namens polymerisierte Kettenreaktion angewandt, die die Art und Weise revolutioniert hat, wie wir DNA sehen und analysieren. 00:33 Die polymerisierte Kettenreaktion ermöglicht es einer Person, bestimmte DNA-Segmente aus einer größeren DNA millionenfach zu amplifizieren. 00:42 Es ist sehr beliebt in forensischen Analysen und wenn Sie jemals eine Krimi-Serie im Fernsehen gesehen haben, haben Sie wahrscheinlich gesehen, wie die PCR in der Praxis angewendet wird. 00:51 Der Prozess der PCR oder polymerisierten Kettenreaktion besteht einfach aus dem Kopieren der Idee der zellulären DNA-Replikation und dies wird dann auf edle Art und Weise genutzt, um um die Replikation der DNA zu bewerkstelligen, die man nur auf eine bestimmte Weise zu kopieren braucht. 01:09 Der Vorteil der PCR ist, dass sie eine sehr einfach durchzuführende Technik ist. 01:13 Ein Highschool-Schüler kann in etwa einer Stunde zur PCR ausgebildet werden. 01:17 Man muss die Sequenz kennen, um damit zu beginnen, und das ist eine der Hauptanforderungen der PCR, wie wir noch sehen werden. 01:25 Die PCR-Technik nutzt nun die Kenntnis der Sequenz, um einen DNA-Primer zu entwerfen und chemisch zu synthetisieren, der die zu amplifizierende Region markieren soll. Das muss ich jetzt erklären. 01:36 In einer früheren Vorlesung habe ich darüber gesprochen, wie die DNA-Polymerase einen Primer verwendet, um die DNA-Replikation zu starten und in der Zelle ist dieser Primer die RNA. 01:46 Das Problem mit dem RNA-Primer ist, dass er entfernt werden muss und dann durch etwas anderes ersetzt werden muss. Das ist ein ziemlich komplizierter Prozess. 01:53 Wenn wir DNA in einem Reagenzglas replizieren wollen und zwar effizient, beginnen wir mit einem DNA-Primer. 01:59 Das Schöne an einem DNA-Primer ist, dass er einen Startpunkt für die Replikation definiert. 02:04 Da ein Primer eine bestimmte Sequenz hat, kann man diese Sequenz entwerfen und dann zulassen, dass die PCR dadurch den richtigen Ort für die Bildung von Basenpaaren findet, und das bestimmt also, wo die Replikation stattfindet, wie wir noch sehen werden. 02:17 Um diesen Prozess in Gang zu setzen, müssen wir eine Ziel-DNA definieren. Diese DNA könnte, sagen wir mal, eine menschliche DNA sein und diese menschliche DNA enthält eine Region, die wir analysieren wollen. 02:28 Vielleicht ist es eine genetische Mutation, die eine Person erlitten hat und wir sind daran interessiert, diese zu bestimmen. 02:33 In jedem Fall würden wir die Sequenzen um die DNA, die wir untersuchen wollen, kennen. Also, sagen wir, wir haben zum Beispiel eine Person, die an Sichelzellenanämie leidet. 02:43 Und bei der Sichelzellenanämie liegt eine Mutation im Hämoglobin-Gen vor. 02:48 Wir kennen die Sequenzen des Hämoglobin-Gens, aber wir kennen nicht die Sequenzen der Mutation in der Mitte des Gens. 02:54 Wir könnten also einen DNA-Primer entwerfen, der zu beiden Enden des Hämoglobin-Gens komplementär ist, und ihn so zu gestalten, dass er nun Basenpaare mit dem Ende des Hämoglobin-Gens in der DNA bildet. 03:08 Wir nehmen die Ziel-DNA, das ist die Person, die die Mutation hat. 03:13 Wir nehmen ihre DNA, mischen sie mit den Primern, mischen sie mit 4 dNTPs und jetzt kommt der Clou, eine thermostabile DNA-Polymerase. Wir werden gleich sehen, warum die thermostabile DNA-Polymerase wichtig ist. Thermostabile bedeutet, dass sie resistent gegen Denaturierung durch Hitze ist. Die meisten Enzyme zerfallen bei Hitze, ein thermostabiles Enzym nicht. 03:39 Wir nehmen dieses ganze System. Diese Mischung, die ich Ihnen gerade definiert habe, und wir fügen es zu einem sogenannten Thermocycling-System hinzu. Und ein Thermocycling-System ist ein Gerät, das diese Probe aufheizen und dann auf verschiedene Temperaturen abkühlen kann, wie wir sehen werden. 03:54 Und damit beginnt ein Prozess, den ich als nächstes beschreiben werde. 03:58 Wir sehen hier also zum Beispiel die Ziel-DNA der Person, die eine mutierte Hämoglobin-Gen trägt. 04:04 Es ist eine Duplex-DNA und ich habe es hier kurz gemacht. 04:09 Aber in Wirklichkeit hätten wir die Gesamtheit der Chromosomen dieser Person. Es gibt hier also eine Menge Sequenzen. 04:15 Der erste Schritt im Prozess der Anwendung der PCR ist das Auseinanderziehen der Stränge. 04:21 Dies ahmt in etwa nach, was die Helikase bei der DNA-Replikation macht, denn hier ziehen wir sie physisch auseinander. 04:28 Aber wie kann man sie auseinanderziehen? Die Art und Weise, wie man man sie auseinanderzieht, ist, dass man sie fast zum Sieden erhitzt. 04:33 Wenn man das tut, werden die Wasserstoffbrücken, die das DNA-Duplex zusammenhalten, gebrochen und die Stränge lösen sich vollständig auseinander. Hier trennen sich also die Stränge. 04:43 Der zweite Teil ist die Abkühlung, sodass die Primer, die in die Lösung gemischt wurden mit ihrer entsprechenden Sequenz Basenpaare bilden können. 04:53 Wenn man nun die Primer in der richtigen Länge herstellt, ist die einzige Stelle, an der sie die Basenpaare bilden werden, dort, wo man sie haben wollte, auf beiden Seite, in diesem Fall des Hämoglobin-Gens. 05:04 Dieser Prozess erfordert eine bestimmte Temperatur, die sogenannte Glühtemperatur. 05:09 Der Thermocycler erhitzt also das System bis zum Siedepunkt und kühlt es dann auf diese magische Temperatur ab, bei der die Primer Basenpaare mit ihren Komplementen bilden. 05:22 Der dritte Schritt besteht dann darin, dass die DNA- Polymerase diese Stränge zu replizieren beginnt. 05:27 Nun, die DNA-Polymerase war bereits in der Mischung und erinnere dich, wir haben eine thermostabile DNA-Polymerase und deshalb wurde sie durch das Kochen nicht beschädigt. 05:39 Die DNA-Polymerase verwendet die 4 dNTPs zur Replikation des Strangs von Interesse und wir können die Replikation, die stattfindet, hier sehen. 05:47 Nachdem das geschehen ist, definieren die Primer die Enden. Ich sehe also ein Ende in gelb und ich sehe ein weiteres Ende in grün. 05:58 Diese Replikation läuft ab und was nun geschieht ist, die Primer starten die Replikation immer und immer wieder, immer und immer wieder die gleichen Stränge. 06:07 Man beachte, dass die beiden Stränge, die wir kopieren, nur der obere und der untere Strang sind. Sie sind eigentlich die gleiche Sequenz. 06:13 Wir erhalten also ein Duplex, das daraus gemacht wurde. 06:16 Das bedeutet, dass wir mit einem Duplex in Schritt 1 starteten. 06:20 Aber in Schritt 3 haben wir zwei Kopien desselben Duplex. 06:25 Aber man kann schon sehen, was hier passiert. Jedes Mal, wenn wir einen Zyklus des Kochens und der Replikation durchführen, verdoppeln wir die Anzahl der Stränge. 06:35 Das mag jetzt nicht wie eine große Sache erscheinen. Aber wenn du 30 Mal machst, hast du 2 hoch 30 mal mehr Stränge als zu Beginn, zumindest theoretisch. Das sind über eine Milliarde. 06:48 Das bedeutet also, dass man aus winzigen Mengen von DNA und unglaubliche Mengen davon mit dieser Methode hervorbringen kann. Deshalb ist es ein sehr mächtiges Werkzeug an einem Tatort. 06:56 Man braucht nicht viel von der DNA eines Verdächtigen, um diese Art von Analyse durchzuführen. 07:02 Dieser Prozess wird in der Regel 30 bis 40 Zyklen lang wiederholt. Es gibt einige Prozesse, bei denen er noch öfter durchgeführt wird. 07:10 Diese leistungsstarke Technik ist eine von vielen, die wir verwenden, um Sequenzen zu analysieren. So erfolgt zum Beispiel die Analyse von DNA durch PCR, aber es gibt auch noch andere Arten von Analysen, die uns interessieren. Und diese Arten von Analysen würde ich gerne zusammenfassen und Omics-Analyse nennen. 07:28 Wir werden gleich sehen, wie das ins Spiel kommt. 07:30 Die technologischen Fortschritte sind es, die die Omics-Analyse erst möglich gemacht haben. 07:36 Omics-Methoden konzentrieren sich nicht auf einzelne Moleküle innerhalb einer Zelle, sondern auf ein breites Spektrum. 07:44 Wenn wir also zum Beispiel über Genomik sprechen, gibt es eine der Omics, die jede DNA-Sequenz in einer Zelle untersuchen. 07:52 Vor 30 Jahren war das noch unvorstellbar, niemand wusste, wie man das macht. 07:57 Jetzt können wir ein Genom in sehr sehr kurzen Zeiträumen sequenzieren, in einer Woche. 08:02 Wir können Transkriptomik betreiben, bei der wir alle Transkripte einer Zelle analysieren. 08:07 Nun sind die Transkripte natürlich die RNAs, die für die Herstellung von Proteinen transkribiert werden. 08:13 Wenn wir alle RNAs einer Zelle kennen und wie viele von ihnen hergestellt werden, haben wir eine erstaunlich umfassende Information darüber, was die Zelle tut und wie viel davon sie zu produzieren versucht. 08:26 Die Proteomik ist ein weiterer Bereich der Omic, der sich mit der Untersuchung der Proteinexpression beschäftigt. 08:33 So wie wir die Transkriptomik nutzen können, um festzustellen wie viele und welche Arten von RNAs gebildet werden, können wir mit der Proteomik feststellen, wie viel von jedem Protein in einer Zelle hergestellt wird. 08:47 Metabolomik ist also eine Analyse des Metaboloms. 08:50 Ein Metabolom entspricht natürlich allen Stoffwechselprodukte, die in einer Zelle gebildet werden. 08:55 Das könnte alle Moleküle umfassen, die in einem Zitronensäurezyklus entstehen und welche Menge von jedem hergestellt wird. Alle Moleküle, die in der Glykolyse hergestellt werden und wie groß die Mengen der einzelnen Moleküle sind. 09:05 Mit anderen Worten, all die verschiedenen biochemischen Prozesse, die sich spektral über die gesamte Zelle abspielen. Das ist der Vorteil von Omics-Typ-Analyse und es gibt Dutzende von Omics-Disziplinen, die die Menschen entwickelten, um diese Art von Analysen durchzuführen. 09:21 Und dadurch sind wir tatsächlich in der Lage, auf Systemebene zu verstehen, was in der Zelle passiert, anstatt nur auf molekularer Ebene zu verstehen, was in der Zelle passiert. 09:33 Es ermöglicht uns, besser zu verstehen, worum es im Leben wirklich geht.
About the Lecture
The lecture DNA Amplification – Analytical Techniques in Biotechnology by Kevin Ahern, PhD is from the course Analytical Techniques in Biotechnology.
Included Quiz Questions
Which of the following is true of the polymerase chain reaction (PCR)?
- It has three steps to each cycle.
- It requires an RNA primer for each round of replication.
- It works best with a heat-sensitive DNA polymerase.
- It typically amplifies DNA about 1000 fold.
- It requires transfer RNA.
Which of the following is true of the steps of the polymerase chain reaction (PCR) process?
- Denaturation is the first step.
- Renaturation must be performed very quickly.
- Elongation must be performed at temperatures close to boiling.
- The number of DNA sequences doubles every 5 cycles.
- Cold solutions are necessary for the reaction.
Which of the following is NOT a component of a PCR reaction mixture?
- RNA primer
- DNA template with target sequence
- Four types of dNTPs
- Taq polymerase
- DNA primers
Which of the following is NOT associated with PCR?
- Denaturation of template DNA — done with the help of thermostable helicase enzyme in the PCR mixture
- DNA primer — determines the replication start point during PCR
- Taq polymerase — a thermostable polymerase enzyme resistant to the denaturation step during PCR
- Thermal cycler — a device used to amplify the target DNA sequence via PCR
- Annealing temperature — temperature which affects the efficiency and specificity of PCR system by facilitating the hybridization of primer to the DNA template
According to the lecture, which of the following best describes the term 'omics'?
- The detection of genes, mRNA, proteins, and metabolites in a specific biological sample
- The replication of target genes in a very short time
- The synthesis of target mRNA with high accuracy and efficiency
- The synthesis of target tRNA and rRNA with high accuracy and efficiency
- The synthesis of the target protein and its related metabolites with high accuracy and efficiency
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
1 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
It is an xcellent explication! I understend it at all. Thank you