Playlist
Show Playlist
Hide Playlist
Antimicrobial Resistance
-
Slides 01 Bacteria MicrobiologyAdvanced.pdf
-
Download Lecture Overview
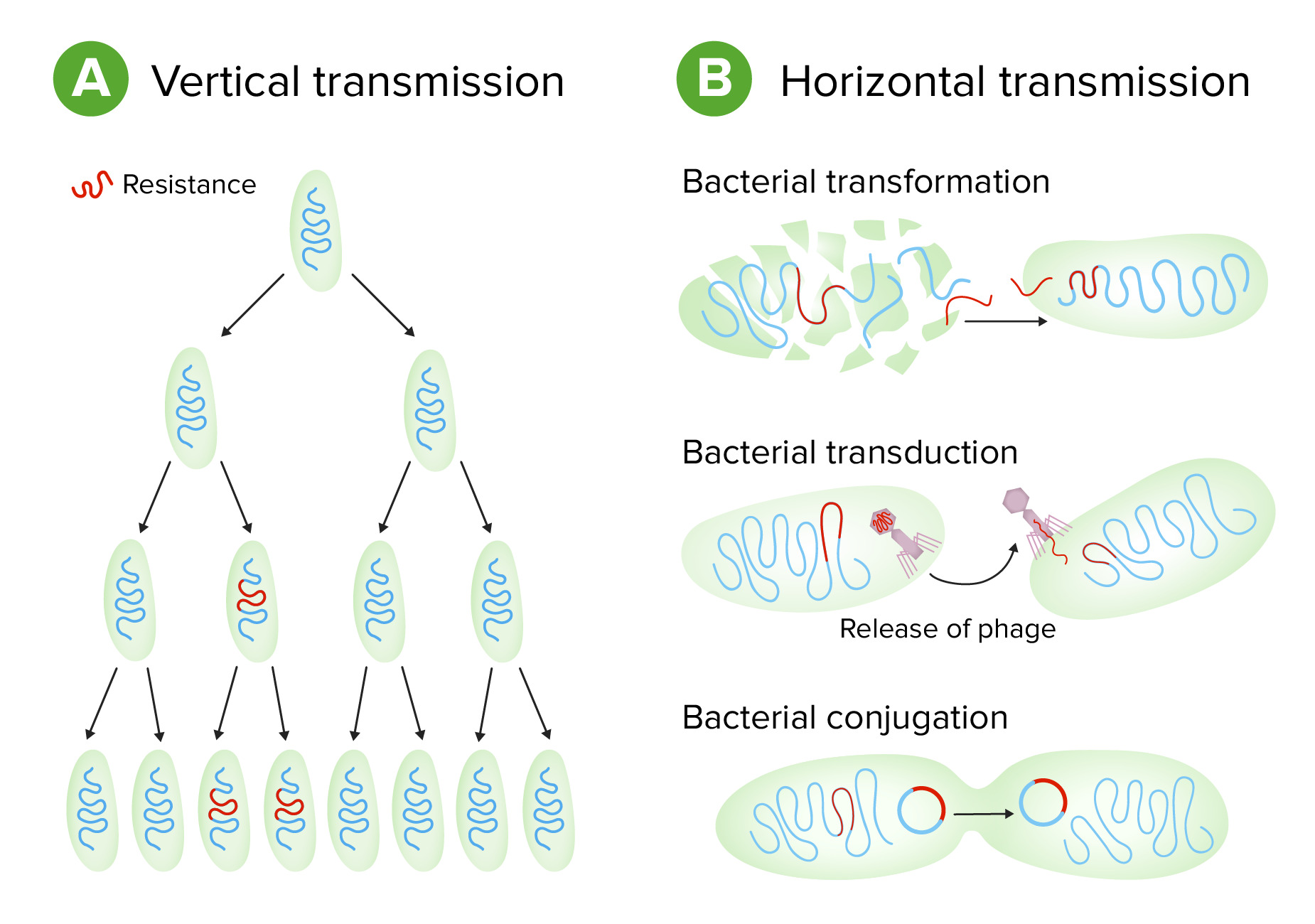
00:01 Leider hat sich durch die Entwicklung neuer antimikrobieller Verbindungen auch schnell eine Resistenz gegen diese entwickelt. 00:11 Und heute, haben wir fast gegen jede antimikrobielle Verbindung, die wir entwickelt haben, eine Resitenz. 00:17 Die Situation wird immer schlimmer, da wir dadurch immer weniger Optionen zur Behandlung von bakterielle Infektionen haben. 00:25 Antimikrobielle Resistenz kommt in der Natur vor und sind uralt. 00:32 Bakterien stellen Antibiotika her, um mit anderen in der Natur zu konkurrieren. 00:35 Viele davon, haben wir uns zur Verwendung als antimikrobielle Behandlung von Infektionen bei Menschen zunutze gemacht. 00:43 Wir wissen, dass es diese Gene, die eine Resistenz verursachen, schon seit Tausenden und Tausende von Jahren gibt. 00:50 Wir können sie in sehr alten Stätten auf der Erde finden und es gibt viele Beweise, dass sie schon existiert haben, lange bevor der Mensch eine eine antimikrobielle Substanz entwickelt hat. 00:59 Wir nutzen also wirklich den Vorteil von etwas, was es schon lange in der Natur gibt. 01:04 Es gibt eine Reihe von Mechanismen, wie diese antimikrobiellen Resistenzgene funktionieren. 01:13 Zum Beispiel, können die Synthese eines Enzyms steuern, das direkt ein Medikament abbaut. 01:18 Dies ist eine einfache Art Widerstand zu leisten. 01:21 Sie können das Medikament chemisch verändern, so dass es also seine Funktion stört. 01:26 Sie könnten die Aufnahme des Medikaments in Zellen und Geweben verhindern, so dass diese sein Ziel nicht mehr erreichen können. 01:32 Oder sie könnten den Export eines Medikamentes aus der Bakterienzelle anregen, so dass es nicht mehr bakterizid ist. 01:40 Oder sie können den Zielort der Medikamente verändern. 01:43 Es gibt also viele verschiedene Mechanismen der antimikrobiellen Resistenz. 01:47 Diese sind alle in Genen kodiert, die für Proteine kodieren, die die diese verschiedenen Aktivitäten haben. 01:55 Lassen Sie uns ein Beispiel nehmen, um das zu veranschaulichen. 01:58 Wir werden als Beispiel das Antibiotikum Vancomycin verwenden. 02:03 Sein Ziel ist die Veränderung der Zellwand. 02:08 Vancomycin wirkt also durch Blockierung des Aufbaus der Murein-Zellwand. 02:14 Oben auf dieser Folie ist die normale Inkorporation der Vorstufen des Mureins. 02:19 Also das blaue und das grüne Oval, das sind Zuckermoleküle, die Teil der wachsende Peptidoglykan-Kette werden. Und diese kleineren Ovale darunter, das sind Aminosäuren, die schließlich das Murein vernetzen, um es sehr stark zu machen. 02:37 Das funktioniert so, dass Untereinheiten zu der wachsenden Kette hinzugefügt werden. 02:41 In der zweiten Zeile dieser Abbildung können Sie die wachsende Polypeptidkette sehen. 02:48 Vancomycin bindet an die Vorläufersubstanz durch Bindung an die Aminosäuren. 02:52 Vancomycin ist hier in lila mit dem V dargestellt, oder vielleicht ist das braun. 02:56 Es bindet die Aminosäuren und blockiert den Einbau von neuen Ketten und hemmt daher die Mureinsynthese und tötet die Bakterien ab. 03:06 Resistenz gegen Vancomycin: Ein Mechanismus der Resistenz ist dass sich das Bakterium einfach verändert. 03:11 Die D-ala-D-ala verändert sich zu D-ala-D-lac, und D-Laktat können in diese Kette eingebaut werden. 03:19 Es umgeht so die Vancomycin-Resistenz und das Antibiotikum wirkt nicht mehr. 03:23 Das ist ein Beispiel dafür, wie Resistenz funktioniert. 03:26 Zurück zu unseren β-Laktam-Antibiotika, die ich bereits erwähnt habe. Der Pfeil zeigt auf den β-Lactamring, die für alle Mitglieder dieser Klasse gelten, weshalb wir sie β-Lactame nennen. 03:40 Wir haben bisher über 300 β-Laktamasen identifiziert. 03:46 Dies sind Enzyme, die den β-Lactam-Ring schneiden. 03:52 Und diese β-Laktamasen kodieren Resistenz zu den β-Laktam-Antibiotika. 03:58 Sie sehen also das Ausmaß des Problems, dass die β-Laktamasen überall sind. 04:03 Erschwerend zu der Antibiotikaresistenz kommt hinzu, dass die Gene, die für Resistenzfaktoren kodieren, die zum Beispiel die β-Laktamasen kodieren, oft in der Lage sind, sich von Bakterium zu Bakterium weiterzugeben. 04:20 Eine Möglichkeit dies zu tun, ist über Plasmide. 04:23 Und in der Tat sind viele dieser Antibiotika-Resistenzgene auf Plasmiden kodiert. 04:28 Die Darstellung, die wir in einer unserer anderen Vorlesungengesehen haben, zeigt, wie sich Plasmide von einer Bakterienzelle zur anderen bewegen. 04:37 Oben links ist eine bakterielle Zelle mit einem Chromosom in grün und einem kleineren Plasmid in rot. 04:43 Nehmen wir an, dieses Plasmid kodiert eine β-Lactamase, das diesem Bakterium eine Resistenz gegen Beta-Lactame verleiht In der zweiten Reihe, tauschen die beiden Bakterien nun die DNA durch einen Pilus, der die beiden Zellen verbindet, aus. 04:56 Und das Plasmid wandert von einer Zelle zur anderen. 04:59 Das Ergebnis ist, dass die zweite Zelle nun Antibiotikaresistenz erworben hat. 05:05 Sie wissen also, dass dies ein Problem ist. 05:07 Wir füttern oft unsere Tiere, die wir als Nahrung zu uns nehmen mit vielen Antibiotika, damit sie schnell heranwachsen. 05:14 Der Effekt ist, dass wir bei den Tieren auf antimikrobielle Resistenz selektieren. 05:18 Und wenn wir dann diese Lebensmittel essen, erwerben wir Antibiotika Resistenzgene in uns, die zunächst keine Rolle spielen. 05:27 Aber wenn wir dann operiert werden und wir eine Antibiotikatherapie brauchen, kann es sein, dass es nicht funktioniert, weil wir die Resistenz bereits in uns haben. 05:35 Diese Antibiotika Resistenzgene können sich ausgiebig in Bakterien bewegen. 05:39 Deshalb sind sie ein Problem. Nicht nur wegen der Plasmidmobilität, aber auch durch Bewegung, durch Transduktion, den Austausch von DNA-Stücken, durch Viren oder einfach durch nackte DNA. 05:52 Gentransfer zwischen Bakterien, wir nennen das horizontalen Gentransfer, ist weit verbreitet und stellt ein großes Problem für die antimikrobielle Resistenz dar. 06:02 Zum Schluß noch eine Darstellung, die einge gängige Mechanismen der Resistenz gegen antimikrobielle Medikamente zeigt. Hier am Beispiel von Penicillinen und Cephalosporine, die durch β-Laktamasen hydrolysiert werden. 06:16 Diese Resistenzgene sind in der Tat auf Plasmiden vorhanden. 06:20 Methicillin-Resistenz ist eine Veränderung des Penicillin-Bindungsprotein, nicht in der β-Lactamase, sondern in einem separaten Protein. 06:27 Dies wird nicht zufällig auf einem Plasmid getragen. 06:30 Tetracyclin-Resistenz kodiert eine Pumpe die den Wirkstoff aus der aus der Bakterienzelle herauspumpt. 06:37 Dies ist ein plasmidbasierter Resistenzfaktor. 06:39 Wenn man sich also all diese verschiedenen Mechanismen der Resistenz Modifikation der Medikamente anschaut, die Synthese von alternativen Substraten, Acetylierung und Veränderung der Bindungsstellen... 06:51 - schau mal wie viele auf Plasmiden kodiert sind. 06:54 Und das bedeutet einfach, daß sie leicht von von einem Bakterium zu einem anderen Bakterium gelangen können und wir eine schwere Zeit vor uns haben, bakterielle Infektionen zu behandeln, wenn diese Resistenzgene so mobil sind.
About the Lecture
The lecture Antimicrobial Resistance by Vincent Racaniello, PhD is from the course Bacteria.
Included Quiz Questions
Vancomycin resistance is mediated by which of the following?
- Genetic mutation resulting in the lack of a vancomycin-binding site
- Increased d-Ala-d-Ala synthesis
- Decreased peptidoglycan wall synthesis
- Decreased bacterial utilization of lactose
- Increased synthesis of the beta-lactam ring
Plasmids containing beta-lactamases would confer resistance against which of the following antibiotics?
- Cephalosporins
- Tetracyclines
- Vancomycin
- Fluoroquinolones
- Linezolid
Which of the following describes the mechanism of resistance to tetracyclines?
- Efflux pump that pushes the drug out of the cell
- Acetylation blocking drug transport into the cell
- Enzymatic modification
- Change in peptidoglycan-binding site
- Beta-lactamases
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
2 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
yes, very good! Very rethorical and good lecture. I love lecturio
short and sweet concepts. nice tables. easy to understand. thank you.