Playlist
Show Playlist
Hide Playlist
Antibody Variable Region Diversity – Lymphocyte Development
-
Slides Adaptive Immune System.pdf
-
Download Lecture Overview
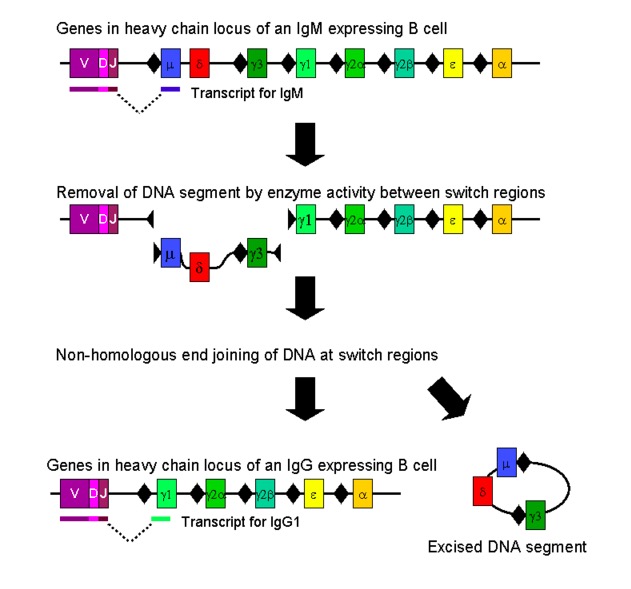
00:01 It's important that B cells are specific for one single antigen, so for example you have one B cell that's specific for staphylococcus, another for streptococcus, another for candida, another for cytomegalovirus, and so on. 00:17 And in order to ensure that the B cell is only specific for a single antigen, you need to just have one heavy chain sequence and one light chain sequence; and that is ensured by a process that is referred to as allelic exclusion. 00:37 The result of allelic exclusion is that if the first attempt at recombining the heavy chain is successful, recombination on the other chain is prevented. 00:49 It's not allowed, so if the first recombination is successful, you make a protein as a feedback mechanism that prevents recombination on the other heavy chain locus. 01:01 Likewise for the light chain recombination, if there's a successful recombination of the light chain, either the kappa light chain or the lambda light chain it prevents subsequent recombination of other light chain loci, so allelic exclusion ensures that there isn't a mixture of different heavy chains or different light chains for a given B cell, just one heavy chain, one light chain. 01:25 There'll be millions of copies but they'll all be identical. 01:28 Why don't we look at the numbers that are involved here, give us an idea about how incredible this mechanism is. 01:36 There are less than 200 genes involved, yet those genes can encode millions of different antibodies. 01:47 So we've already heard that there are around about 40 variable gene segments for the immunoglobulin heavy chain, 27 diversity gene segments, and half a dozen or so joining gene segments. 02:00 And we've heard that this recombination is a random process, so given B cell can pick anyone out of the 40, anyone out of the 27, anyone out of the 6 for the V, D and J. So because any of those can be picked, you can multiply those three numbers by each other, 40 times 27 times 6, so that gives us nearly six and a half thousand different possible sequences, protein sequences, from whatever we got there, 40 plus 27 that's 67 plus 6, 73 if my math is right, okay, so a very small number genes producing thousands of different protein sequences. 02:49 And for the kappa light chain -- 40 variable gene segments, anyone of which can be chosen, no D remember in the light chain, but 5 J so we can multiply 40 by 5 that gives us 200, that's quite a simple solving to do and in a given B cell, it may make exactly the same heavy chain rearrangement as another B cell, but a different light chain, so we can actually take those two numbers and multiply them together 6,480 times 200 is I'm sure you already worked out by mental arithmetic, is 1.3 times 76, 1.3 million different protein sequences from that way under 200 different gene segments. 03:36 And of course, there's also the lambda light chain -- 30 variable gene segments, 5 joining gene segments gives us a 150, that gives us a figure of nearly a million different sequences, you can add those two together, so over two million different protein sequences from less than 200 genes. 03:56 but that's only the start of it, the antibodies can have much more variability than that. 04:02 There are a variety of mechanisms that create additional variability. 04:08 Maybe you're wondering where this additional variability comes from and essentially there are three mechanisms that create variability that goes way beyond what occurs due to the recombination of the V, D, and J in the heavy chain and V and J in the light chain. 04:27 First of all, when that recombination process occurs, it's not an absolutely set point at which the splicing of D to J and then V to D J in the heavy chain and V to J in the light chain occurs. 04:43 In other words there are recombinatorial inaccuracies. 04:47 Now this isn't always a good thing, this is one of the ways in which for example a stop code on might be inserted and you're not able to make an antibody molecule, but it does greatly increase the diversity and that's beneficial because there are lots and lots of different pathogens out there and they're mutating all the time so we need to have an enormous diversity of antibody molecules and T cell receptor molecules. 05:14 Secondly, there is a process referred to as N-nucleotide addition and the N stands for nontemplated and this is a mechanism that as we'll see is mediated by an enzyme, terminal deoxynucleotidyl transferase. 05:32 I've already mentioned that enzyme a little bit earlier in this lecture -- terminal deoxynucleotidyl transferase or TdT, mediasis and nucleotide addition. 05:43 And then, finally, somatic hypermutation. 05:47 Now, all genes mutate. But in the immunoglobulin genes, one observes a much higher rate of mutation than in other genes, at a thousand times more mutations are permitted within the immunoglobulin genes by a variety of mechanisms and this means that the diversity of antibodies can be increased enormously.
About the Lecture
The lecture Antibody Variable Region Diversity – Lymphocyte Development by Peter Delves, PhD is from the course Adaptive Immune System. It contains the following chapters:
- Allelic Exclusion
- How Many Different Antibodies are Possible?
- Overview of the Additional Variability of Immunoglobulin Genes
Included Quiz Questions
Which of the following is NOT considered a main cause of antibody diversity?
- Recombination signal sequences
- Recombination
- Somatic hypermutation
- N-nucleotide addition
- Recombinatorial inaccuracies
The addition of nontemplated nucleotides is mediated by which of the following?
- Terminal deoxynucleotidyl transferase
- Immunoglobulin heavy chain
- Somatic hypermutation
- Light chain gene segments
- Recombination signal sequences
Customer reviews
5,0 of 5 stars
| 5 Stars |
|
1 |
| 4 Stars |
|
0 |
| 3 Stars |
|
0 |
| 2 Stars |
|
0 |
| 1 Star |
|
0 |
Reading a textbook could be boring. The lecture nailed what I need to know.